Molecular mechanisms of pathological tumor transformation and their clinical implications: predictors of pulmonary adenocarcinoma transformation into small cell carcinoma
Lee et al. have recently described predictors of transformation into small-cell carcinomas from lung adenocarcinomas under treatment by tyrosine kinase inhibitor (1). Their very interesting work raises several reflections about tumor transformation and the clinical management of tumor drug resistance.
The occurrence of a less differentiated component is described in the course of various neoplasms (Table 1). The genetic hallmark of the primary tumor is kept in the transformed component. Dedifferentiated liposarcomas generally display more extensive chromosomal abnormalities than well differentiated liposarcomas. However, both tumors exhibit the same MDM2 and CDK4 amplifications (2). Most high-grade lymphomas occurring during the course of chronic lymphoid leukemia are clonally related to the preceding disease, thus representing true transformation. Chronic lymphoid leukemia harboring NOTCH1 mutations, or expressing a stereotyped B-cell receptor (BCR) belonging to subset 8, have a significantly higher probability of developing Richter’s syndrome. Chronic lymphoid leukemia treatments (including Bruton’s tyrosine kinase inhibitors) do not seem to affect the risk of transformation. TP53 and MYC abnormalities represent the most frequent genetic lesions and are generally acquired at the time of lymphomatous transformation (3). Blast crisis occurring in the course of chronic myeloid leukemia is associated with additional cytogenetic aberrations to the Philadelphia (Ph) chromosome. A variety of mutations has been associated with progression to blast crisis, especially BCR-ABL tyrosine kinase domain, p53 and p16 mutations (4). The interesting point in this case is that blast crisis can be prevented by an efficient treatment of the chronic disease and elimination of BCR-ABL transcript. Incidentally, a blast crisis has been reported in a patient with chronic myeloid leukemia, after initiation of a treatment with tyrosine kinase inhibitor for EGFR mutated lung adenocarcinomas (5). It is noteworthy that both lymphoid and myeloid chronic leukemia are also tyrosine kinase inhibitors sensitive. Molecular mechanisms for tumor transformation are still not completely understood. These examples show that they appear not unique but depend on tumor type. This underlines the importance of studies like that performed by Lee and coll. for EGFR mutated lung adenocarcinomas.
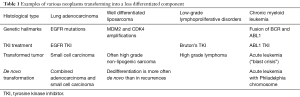
Full table
Over the last decades, the incidence of lung adenocarcinoma has raised worldwide to the point that it represents the most common histological type of lung cancer (6). The advances in the understanding of lung tumor biology have disclosed major cancer gene drivers such as EGFR, KRAS, BRAF, PIK3K, ALK, ROS1, RET, HER2, MEK, MET (7-9), which represent, at least in part, the target of innovative therapies. As a consequence, the EGFR inhibitors (erlotinib, gefitinib, afatinib), have now been introduced in the clinical practice of lung cancer harboring EGFR activating mutations and constitute an effective first-line therapy. Nevertheless, along the treatment, cancer cells acquire a resistant phenotype, through different mechanisms, consisting either in the gain of new EGFR mutations or in the acquisition of other oncogenic drivers bypassing EGFR signaling (10,11). Thr790Met mutation in EGFR gene enhances the affinity of the EGFR for ATP, leading to the continuous activation of EGFR signaling even in presence of the inhibitor (12). The acquisition of T790M represents the most common molecular mechanism of TKI resistance in EGFR mutated adenocarcinoma, occurring in half of the resistant tumors (12). The development of PIK3CA mutations has also been identified as a mechanism of EGFR drug resistance (13) in 5% of cases. MET or HER2 amplification constitute two additional biological mechanisms inducing EGFR inhibitor resistance, accounting for about 15–20% of cases (10,14).
The histological analysis of tissue specimen from patients having developed an EGFR inhibitor resistant tumor has disclosed significant histological modifications, leading to the description of an additional mechanism of drug resistance: the transformation of adenocarcinoma to small cell lung cancer (15). This process, initially, described in case reports (16), has been increasingly characterized in case series, in which repeat biopsies were performed in patients with acquired resistance to EGFR inhibitors. The prevalence of this mechanism of resistance is difficult to evaluate: in a large case series, SCLC was identified in 14% of patients with EGFR mutated adenocarcinoma (17). The genomic analysis of SCLC developed in this setting has disclosed that the EGFR mutations originally detected in adenocarcinoma were retained in SCLC, together with classic oncogenetic drivers such as RB1, TP53 and MYC. This finding has raised the question about SCLC origin: are SCLCs the result of EGFR inhibitors treatment or are EGFR mutated adenocarcinomas more likely to evolve toward SCLC? It has been previously reported that the development of small cell lung carcinomas in association with pulmonary adenocarcinoma, either synchronously or metachronously, could be linked to EGFR mutations, regardless of tyrosine kinase inhibitors use (15). The description of synchronous adenocarcinoma and SCLC before the administration of EGFR inhibitors has suggested that SCLC are not the mere consequence of EGFR inhibitors. Moreover, the progression of NSCLC to SCLC has been documented before the introduction of EGFR inhibitors, with a reported frequency of 5% in a case series (18).
The biological mechanisms underlying the transformation of adenocarcinoma into SCLC are largely unknown. The hypothesis about the existence of a “pool” of pluripotent cancer cell, giving rise to a phenotypically distinct cancer population, constitutes an attractive biological mechanism potentially related with cancer cell therapeutic resistance (19,20).
Moreover, in the field of pulmonary oncology, the hypothesis of the existence of common cells of origin for lung adenocarcinoma and SCLC is gaining ground, since studies in animal models of lung cancer have provided insights about the capacity of alveolar type II cells to give rise to both SCLC and EGFR-mutated adenocarcinoma (21-23).
The article by Lee et al. provides important advancements about the pathogenesis of such mechanism of EGFR-inhibitor resistance (1). Through the genomic analysis of repeat biopsy of patients diagnosed with EGFR-mutated lung adenocarcinoma, the authors found that the alterations of the cancer gene drivers RB1 and TP53 are present even in early stage lung adenocarcinoma. Moreover, such alterations would be correlated with the risk of SCLC transformation after EGFR-inhibitors treatment. Those findings strongly support the hypothesis that SCLC is derived from divergent clonal evolution occurring in early stage lung adenocarcinoma.
Otherwise, Lee et al. are making the very pertinent suggestion that induced hypermutation could occur in latent cancer cells and could be reminiscent of stress-induced hypermutation related to phenotypic switch in microbes during antimicrobial treatment, which maximizes their adaptability against selection pressure from the drugs (1). If this hypothesis proved to be true or if some types of tumor drug resistance were similar to drug microorganisms’ resistance, one could imagine fighting against tumor resistance in a similar way than against microorganism resistance. One example could be the use of associated drugs such as multiple targeted therapies against various tumor genetic mutations as for combined therapy against tuberculosis or against Human Immunodeficiency virus.
Eventually, an important finding, carrying potential implication in pathology practice is the possibility to screen lung adenocarcinomas at risk of SCLC transformation by testing RB1 and TP53 expression in cancer tissue (1). Indeed, genetic analysis of those genes is particularly laborious and requires specific skills in genotyping, since mutations are distributed along the gene length. Testing the expression of Rb and Tp53 in cancer tissue specimens by IHC seems to be well correlated with the inactivation of the respective genes and can be routinely performed in pathology facility. Indeed, characterization of tumors, with numerous biomarkers and genetic abnormalities, is becoming more and more complex. However, rapid initiation of appropriate treatments in clinical practice requires molecular and pathological diagnoses with short delays. Long and complex techniques used for research are likely to be replaced by simple and rapid ones for current practice over time. For example, this is especially true for the assessment of lung carcinoma EGFR and KRAS mutational status, which can presently be obtained in few hours with easy to use techniques (24).
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Lee JK, Lee J, Kim S, et al. Clonal history and genetic predictors of transformation into small-cell carcinomas from lung adenocarcinomas. J Clin Oncol 2017;35:3065-74. [PubMed]
- Asano N, Yoshida A, Mitani S, et al. Frequent amplification of receptor tyrosine kinase genes in welldifferentiated/ dedifferentiated liposarcoma. Oncotarget 2017;8:12941-52. [PubMed]
- Rossi D. Richter's syndrome: Novel and promising therapeutic alternatives. Best Pract Res Clin Haematol 2016;29:30-9. [Crossref] [PubMed]
- Hehlmann R, Saußele S, Voskanyan A, et al. Management of CML-blast crisis. Best Pract Res Clin Haematol 2016;29:295-307. [Crossref] [PubMed]
- Ogata H, Okamoto I, Yoshimoto G, et al. Chronic myelomonocytic leukemia blast crisis in a patient with advanced non-small cell lung cancer treated with EGFR tyrosine kinase inhibitors. Respir Investig 2017;55:181-3. [Crossref] [PubMed]
- Houston KA, Henley SJ, Li J, et al. Patterns in lung cancer incidence rates and trends by histologic type in the United States, 2004-2009. Lung Cancer 2014;86:22-8. [Crossref] [PubMed]
- Kempf E, Rousseau B, Besse B, et al. KRAS oncogene in lung cancer: focus on molecularly driven clinical trials. Eur Respir Rev 2016;25:71-6. [Crossref] [PubMed]
- Wallerek S, Sørensen JB. Biomarkers for efficacy of adjuvant chemotherapy following complete resection in NSCLC stages I-IIIA. Eur Respir Rev 2015;24:340-55. [Crossref] [PubMed]
- Sculier JP, Berghmans T, Meert AP. Advances in target therapy in lung cancer. Eur Respir Rev 2015;24:23-9. [Crossref] [PubMed]
- Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26-6. [Crossref] [PubMed]
- Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 2013;19:2240-7. [Crossref] [PubMed]
- Yun CH, Mengwasser KE, Toms AV, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A 2008;105:2070-5. [Crossref] [PubMed]
- Engelman JA, Mukohara T, Zejnullahu K, et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest 2006;116:2695-706. [Crossref] [PubMed]
- Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007;316:1039-43. [Crossref] [PubMed]
- Norkowski E, Ghigna MR, Lacroix L, et al. Small-cell carcinoma in the setting of pulmonary adenocarcinoma: new insights in the era of molecular pathology. J Thorac Oncol 2013;8:1265-71. [Crossref] [PubMed]
- Zakowski MF, Ladanyi M, Kris MG. Memorial Sloan-Kettering Cancer Center Lung Cancer OncoGenome Group. EGFR mutations in small-cell lung cancers in patients who have never smoked. N Engl J Med 2006;355:213-5. [Crossref] [PubMed]
- Fukui T, Tsuta K, Furuta K, et al. Epidermal growth factor receptor mutation status and clinicopathological features of combined small cell carcinoma with adenocarcinoma of the lung. Cancer Sci 2007;98:1714-9. [Crossref] [PubMed]
- Adelstein DJ, Tomashefski JF, Snow NJ, et al. Mixed small cell and non-small cell lung cancer. Chest 1986;89:699-704. [Crossref] [PubMed]
- Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer 2008;8:755-68. [Crossref] [PubMed]
- Rosen JM, Jordan CT. The increasing complexity of the cancer stem cell paradigm. Science 2009;324:1670-3. [Crossref] [PubMed]
- Sutherland KD, Proost N, Brouns I, et al. Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell 2011;19:754-64. [Crossref] [PubMed]
- Mainardi S, Mijimolle N, Francoz S, et al. Identification of cancer initiating cells in K-Ras driven lung adenocarcinoma. Proc Natl Acad Sci U S A 2014;111:255-60. [Crossref] [PubMed]
- Sutherland KD, Song JY, Kwon MC, et al. Multiple cells-of-origin of mutant K-Ras-induced mouse lung adenocarcinoma. Proc Natl Acad Sci U S A 2014;111:4952-7. [Crossref] [PubMed]
- Thomas De Montpréville V, Ghigna MR, et al. EGFR and KRAS molecular genotyping for pulmonary carcinomas: Feasibility of a simple and rapid technique implementable in any department of pathology. Pathol Res Pract 2017;213:793-8. [Crossref] [PubMed]


