Optogenetic tools for cell biological applications
In the past decade, light-responsive proteins have emerged as impactful tools for investigating a range of biological functions, from controlling neuronal functions to activating or repressing gene expression and triggering signal transduction. In contrast to traditional approaches for controlling cell signaling, such as gene overexpression or the use of soluble agonists, the field of optogenetics uses light-responsive proteins to control the activity of a desired signaling protein. The advantage of using light is that the timing, location, and intensity of the signal can be dynamically and precisely controlled using optics (1). Furthermore, unlike small molecules or purified proteins used in cell signaling studies, optogenetic techniques have reduced off-target effects and are relatively inexpensive, as light can be easily generated by a light source such as a light-emitting diode (LED) or laser.
Early optogenetic studies engineered light-responsive ion channels (i.e., members of the microbial opsin family of proteins) to study the mammalian nervous system (2). The subsequent development of various non-opsin based optogenetic systems has allowed researchers to use light-sensitive protein-protein interactions to modulate cellular activities (1). The tools in this category can be broadly categorized into dimerizers and oligomerizers. Of the dimerizers, the most popular ones include heterodimerizers such as the cryptochrome2 (Cry2/Cry2PHR) and CIB1/CIBN protein pair and the phytochrome B (PhyB) and PIF protein pair, as well as the homodimerizer Vivid (VVD), a small light-oxygen-voltage (LOV) domain containing protein. As a particularly promising system, the Cry2-CIB1 (Cry2PHR-CIBN) system undergoes robust and fast heterodimerization in the presence of blue light (488 nm) and dissociates slowly when the light is switched off (3). The salient advantage of this system is that it does not require the addition of an exogenous chromophore and the slow dissociation of the protein pairs allow for protein-protein interactions for an extended period. Bugaj et al. have also used the homo-oligomerizing properties of Cry2 to activate the canonical Wnt signaling and Rho GTPase signaling cascades (4). Also, Taslimi et al. showed that an E490G mutation in the Cry2 protein enhanced the extent of clustering in this system (5). In contrast to the blue light activated Cry2, the Phy/PIF system responds to red (650 nm) and far-red (750 nm) illumination. Phytochrome B (PhyB) binds to phytochrome-interacting factor 6 (PIF6) in the presence of red-light and dissociates from PIF6 when exposed to far-red (750 nm) light (6).
The current optogenetic systems are not without limitations, and a significant amount of work is currently being invested to improve the properties of these tools. The key drawback of the Cry2PHR/CIBN system is that phototoxicity of blue light can limit its application in vivo. The alternative Phy/PIF system exhibits high reversibility and speed of dissociation; however, one of its key limitations is the requirement of adding an exogenous chromophore without which the system cannot respond to light in mammalian cells. Keeping this in mind, a near-infrared (NIR)-responsive dimerizing system was engineered consisting of the bacterial phytochrome BphP1 (whose chromophore is endogenously present in mammalian cells) and its partner PpsR2 (7) (Figure 1A). In a recent study, Redchuk et al. have addressed some of the limitations of this BphP1-PpsR2 system, e.g., the large size of PpsR2 and its propensity to form oligomers and thereby reduce its efficiency for dimer formation with BphP1 (8).
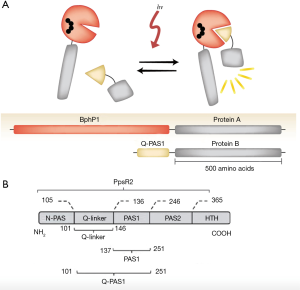
Specifically, Redchuk et al. designed a truncated version of PpsR2 (50 kDa in size), called Q-PAS1 (17 kDa in size), by eliminating the domains involved in the oligomerization of PpsR2 (Figure 1B). The BphP1-Q-PAS1 system exhibited robust and reversible interaction upon illumination with NIR (740–780 nm) light. When used for activating transcription in the nucleus, the BphP1-Q-PAS1 dimerizer system showed faster activation properties compared to the BphP1-PpsR2 counterpart, presumably because of faster transport of Q-PAS1 to the nucleus owing to its smaller size and lack of oligomerization. Due to the smaller size of the Q-PAS1 domain, the BphP1-Q-PAS1 system can also be delivered to mammalian cells on a single DNA vector, simplifying viral gene delivery strategies and ensuring a defined stoichiometric ratio of the two dimerizing domains. The BphP1-Q-PAS1 system was shown to activate and repress gene transcription in mammalian cells as well as to alter the chromatin epigenetic state by recruiting the mSin3-HDAC2 (histone deacetylase 2) complex to the gene of interest in a NIR-light dependent manner. This recruitment resulted in the downregulation of the transcription of the target gene via histone deacetylation and the formation of heterochromatin.
The BphP1-Q-PAS1 system is particularly attractive because of its activation with NIR light, making it unique among other dimerizing systems (which respond to blue or red light). NIR light is less phototoxic to cells, an advantage for in vitro studies using cell cultures, and has deeper tissue penetration, allowing more extensive in vivo studies in animal models. Perhaps the most unique feature of the BphP1-Q-PAS1 system is its lack of spectral cross-talk with blue-light responsive systems. By using the above system in conjunction with the blue-light responsive optogenetic protein-caging system, AsLOV2 (light-oxygen-voltage domain of Avena sativa phototropin 1), the researchers demonstrated tridirectional translocation (membrane, cytoplasm and nucleus) of intracellular proteins (Figure 2). Such spectral multiplexing allows for intricate studies of how multiple pathways interact to govern cell behavior, further expanding the optogenetic toolbox.
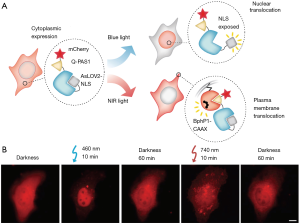
Improving the properties of optogenetic systems is an important step in making optogenetics a robust tool for biomedical research, and some limitations remain to be overcome. Further decreasing the size of the optogenetic domains is particularly important for large proteins like BphP1, which is nearly 5 times larger than Q-PAS1. Protein engineering strategies can also be applied to improving the dynamic range and reducing the dark state activity of the dimerizing domains. Another important consideration for biological experiments is to determine optimal light intensity for highest signal activation without phototoxicity and to optimize protein expression levels to ensure that overexpression of exogenous proteins has no unintended effects on cell behavior.
Optogenetic tools have already been applied to the fields of cancer biology, neurobiology, and synthetic biology, and optimization of the technology further expands the possible applications of optogenetics. Key signaling pathways involved in cell proliferation, tumorigenesis and inflammation (e.g., the Ras/Raf/Mek/Erk cascade, Wnt pathway, PI3K/Akt pathway) have been optogenetically controlled in numerous cell types (1). Many more pathways can be controlled by engineering new optogenetic fusions with receptors or signaling domains of interest, activating or de-activating a pathway at a user-defined node in the signaling cascade. Optogenetic proteins can also be fused with specialized protein domains that enable control of transcription activation, chromatin epigenetic state, or genome modification using light. In this manner, cell processes such as growth, fibrosis, inflammation, migration, metabolism, and morphogenesis can be controlled with the spatial and temporal precision of optogenetics. Optogenetics can also be used as a research tool in vivo, where external illumination on the skin surface or implanted LEDs activate cell behavior in tissues of interest. As a proof of principle, Wang et al. controlled blood glucose levels in mice using external illumination of optogenetic proteins expressed in the liver (9). Such approaches can be extended to many other organs and biological processes, particularly in light of recent engineering advances in minimally invasive, implantable LED devices (1).
With the development of a diverse set of light-responsive systems and illumination technologies, the field of optogenetics has enormous potential for novel findings in how cells and tissues respond to dynamic, spatially and temporally varying signals. Optogenetics is now well poised to become a powerful tool for biomedical research and shed light on signaling dynamics both in vitro and in vivo.
Acknowledgements
Funding: DV Schaffer and RS Kane acknowledge support from NIH grants R01NS087253 and R01NS083856.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Repina NA, Rosenbloom A, Mukherjee A, et al. At light speed: advances in optogenetic systems for regulating cell signaling and behavior. Annu Rev Chem Biomol Eng 2017;8:13-39. [Crossref] [PubMed]
- Optogenetics Deisseroth K. Nat Methods 2011;8:26-9. [Crossref] [PubMed]
- Kennedy MJ, Hughes RM, Peteya LA, et al. Rapid blue light induction of protein interactions in living cells. Nat Methods 2010;7:973-5. [Crossref] [PubMed]
- Bugaj LJ, Choksi AT, Mesuda CK, et al. Optogenetic protein clustering and signaling activation in mammalian cells. Nat Methods 2013;10:249-52. [Crossref] [PubMed]
- Taslimi A, Vrana JD, Chen D, et al. An optimized optogenetic clustering tool for probing protein interaction and function. Nat Commun 2014;5:4925. [Crossref] [PubMed]
- Levskaya A, Weiner OD, Lim WA, et al. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 2009;461:997-1001. [Crossref] [PubMed]
- Kaberniuk AA, Shemetov AA, Verkhusha VV. A bacterial phytochrome-based optogenetic system controllable with near-infrared light. Nat Methods 2016;13:591-7. [Crossref] [PubMed]
- Redchuk TA, Omelina ES, Chernov KG, et al. Near-infrared optogenetic pair for protein regulation and spectral multiplexing. Nat Chem Biol 2017;13:633-9. [Crossref] [PubMed]
- Wang X, Chen X, Yang Y. Spatiotemporal control of gene expression by a light-switchable transgene system. Nat Methods 2012;9:266-9. [Crossref] [PubMed]


