PNUTS at the crossroads of tumorigenesis and metastasis formation
Around 75% of the human genome is transcribed (1,2). Of these genomic transcripts, at most 2% encode functional proteins within their open reading frames. The remaining 98% are non-coding RNAs that lack open reading frames. This majority of RNAs consists to 20% of short non-coding RNAs. These are 20–200 nt in length and include the subgroup of micro RNAs (miRNAs). The remaining 80% of the non-coding RNAs are long non-coding RNAs (lncRNAs), which are 200 nt to 10 kb in length (Figure 1).
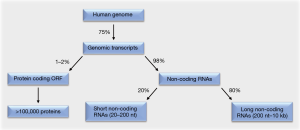
While initially believed to be “junk” RNA, increasing evidence illustrates that non-coding RNAs can be critical regulators of gene and protein expression and hence key regulators of cell fate decisions (1-4). Since these decisions include all steps of cellular transformation and tumorigenesis (2), analyses to comprehensively understand pro- and anti-oncogenic functions of these regulatory RNA molecules are warranted.
Grelet and colleagues have elucidated that a splice switch in the PNUTS/PPP1R10 RNA is relevant for metastasis-associated gene expression programs and tumor growth (5). The PNUTS pre-RNA can give rise to the PNUTS mRNA or an alternatively spliced isoform that represents a tumor-relevant lncRNA of PNUTS (lncRNA PNUTS) (Figure 2). This lncRNA is generated through a removal of 61 bases in the 5'-region of exon 12, leading to a frameshift in the transcript’s open reading frame. As the lncRNA PNUTS is not translated into a protein and only occurs in the monosomal fraction of ribosomes, it is truly non-coding (5). This is of importance in light of the fact that the PNUTS protein, which is also known as serine/threonine phosphatase-1 regulatory subunit-10, contributes to cancer cell migration in conjunction with the lncRNA BCAR4 (6).
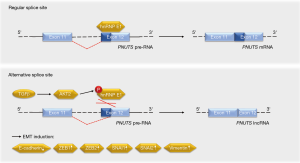
How did Grelet and colleagues identify lncRNA PNUTS as a regulator of the metastatic process? Based on their previous finding that the heterogeneous nuclear ribonucleoprotein E1 (hnRNP E1) controls oncologically important RNA processing (7,8), the authors knocked down hnRNP E1 in tumorigenic epithelial murine NMuMG breast cells and performed a RNA microarray analysis. Data collected with this approach suggested that the knockdown of hnRNP E1 led to an increase in lncRNA PNUTS. A knockdown of hnRNP E1 in epithelial human A549 lung carcinoma cells verified the enhanced processing of PNUTS pre-RNA into lncRNA PNUTS instead of PNUTS mRNA in the absence of hnRNP E1 (5). This finding is consistent with the ability of hnRNP E1 to control alternative splicing (9,10).
Grelet et al. provide several lines of evidence that suggest a tumorigenic role for lncRNA PNUTS. First, the expression levels of lncRNA PNUTS are higher in human breast tumor tissue compared to normal breast tissue. Furthermore, the mesenchymal human MDA-231 breast adenocarcinoma cell line and two lung and bone metastases thereof express higher amounts of lncRNA PNUTS than epithelial breast tumor cell lines. This difference in lncRNA PNUTS expression correlates with a decrease in the levels of the epithelial marker E-cadherin, an increase in the mesenchymal marker vimentin, and higher levels of the transcription factors ZEB1 and ZEB2 (5). Together with other transcription factors, ZEB1 and ZEB2 promote the epithelial-to-mesenchymal transition (EMT). This essential developmental process is hijacked by cancer cells during tumorigenesis to develop tumor metastases at distant sites (11-13). Overexpression of the lncRNA PNUTS in A549 and NMuMG cells verified that this lncRNA promotes the accumulation of mesenchymal marker proteins together with an increased migratory and invasive potential (5).
At the molecular level, hnRNP E1 binds to exon 12 of the PNUTS pre-RNA to control its splicing (5). Stimulation with the EMT-promoting cytokine TGFβ activates the kinase AKT2, which phosphorylates hnRNP E1 and thereby disrupts its interaction with the PNUTS pre-RNA (Figure 2). As this effect occurs transiently, it may rather contribute to the initiation of TGFβ-induced EMT than to the progression of the metastatic cascade. On the other hand, the constantly elevated levels of lncRNA PNUTS in tumors and mesenchymal cell lines may indicate additional functions of this molecule. It will be interesting to see in this context whether the frequently observed overexpression and hyperactivation of AKT2, which correlates positively with the aggressiveness of breast and other cancers (14), is linked to the ratio of PNUTS RNA species.
How does the lncRNA PNUTS regulate EMT? The lncRNA PNUTS acts as a competing endogenous RNA sponge for the miRNA miR-205 (5), which has a significant impact on EMT (15). Although the lncRNA PNUTS can bind and inactivate six miR-205 molecules, the PNUTS mRNA does not sequester this miRNA. Structural features and ribosomal hindrance are discussed as potential mechanisms underlying this finding (5).
The formation of metastases in vivo is a complex process that involves a plethora of factors. Grelet and colleagues analyzed the impact of lncRNA PNUTS in a mammary fat pad transplantation model using NOD/SCID mice and epithelial MDA-468 cells or mesenchymal MDA-231 lung metastasis cells. They could show that an overexpression of lncRNA PNUTS in MDA-468 cells increased and that an attenuation of lncRNA PNUTS expression in MDA-231 cells decreased tumor growth, respectively. The authors also show that lncRNA PNUTS levels determine the primary growth of MDA-231 cells in fat pads (5). Therefore, it will be interesting to see the relative contributions of altered EMT signaling and hitherto unknown effects of the lncRNA PNUTS on tumor cell growth. It is equally thought-provoking why MDA-231 breast cancer cells and metastases thereof have equal levels of the lncRNA PNUTS but express divergent levels of epithelial and mesenchymal marker proteins. Answers to these questions might contribute to a better understanding of the complex regulatory mechanisms involved in the process of metastatic cancer cell growth.
The identification of lncRNA PNUTS as critical modulator of EMT supports the view that EMT is a main driver of the metastatic spread of breast cancer cells (16). At present, it is also possible that lncRNA PNUTS expression is causally associated with a decreased chemosensitivity of mesenchymal cancer cells (17,18). Another crucial question is whether there are further upstream regulators of the PNUTS lncRNA/mRNA splice switch and if these might represent feasible drug targets. There could be additional mechanisms beyond TGFβ and AKT signaling; for example, a possible interplay between epigenetic modifiers and RNA-dependent tumor-relevant mechanisms (3,19). One may also consider that further miRNAs might be competed out by the lncRNA PNUTS (5). Since the miR-205 itself can exert pro- and anti-oncogenic functions in an ambivalent manner (15), additional work is necessary to generate safe and effective RNA-based therapeutics. Such drugs may pave the way for innovative and effective treatment strategies against diverse cancer types. Furthermore, as elevated lncRNA PNUTS expression is an early event in TGFβ-mediated EMT (5), it could be an early prognostic marker for EMT-mediated drug resistance (17,18).
Taken together, in their work Grelet et al. (5), the team around Philip H. Howe and colleagues has revealed for the first time that bifunctional RNA isoforms are able to dynamically regulate biologically important cellular signaling pathways. Moreover, as these authors demonstrate that TGFβ and AKT signaling trigger the alternative splicing of the PNUTS pre-RNA, the expression levels of the lncRNA PNUTS might serve as a biomarker to define patient subgroups, whose primary tumors are prone to metastasis formation. These patients might particularly benefit from targeted therapies that abrogate TGFβ signaling and AKT activation (14,20).
Acknowledgements
Funding: This work was supported by the Wilhelm Sander-Stiftung (grant number, #2010.078).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Serviss JT, Johnsson P, Grander D. An emerging role for long non-coding RNAs in cancer metastasis. Front Genet 2014;5:234. [Crossref] [PubMed]
- Shi T, Gao G, Cao Y. Long Noncoding RNAs as Novel Biomarkers Have a Promising Future in Cancer Diagnostics. Dis Markers 2016;2016:9085195.
- Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol 2013;10:925-33. [Crossref] [PubMed]
- Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017;16:203-22. [Crossref] [PubMed]
- Grelet S, Link LA, Howley B, et al. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat Cell Biol 2017;19:1105-15. [Crossref] [PubMed]
- Xing Z, Lin A, Li C, et al. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell 2014;159:1110-25. [Crossref] [PubMed]
- Chaudhury A, Hussey GS, Ray PS, et al. TGF-beta-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat Cell Biol 2010;12:286-93. [PubMed]
- Hussey GS, Chaudhury A, Dawson AE, et al. Identification of an mRNP complex regulating tumorigenesis at the translational elongation step. Mol Cell 2011;41:419-31. [Crossref] [PubMed]
- Zhang T, Huang XH, Dong L, et al. PCBP-1 regulates alternative splicing of the CD44 gene and inhibits invasion in human hepatoma cell line HepG2 cells. Mol Cancer 2010;9:72. [Crossref] [PubMed]
- Akker SA, Misra S, Aslam S, et al. Pre-spliceosomal binding of U1 small nuclear ribonucleoprotein (RNP) and heterogenous nuclear RNP E1 is associated with suppression of a growth hormone receptor pseudoexon. Mol Endocrinol 2007;21:2529-40. [Crossref] [PubMed]
- Brabletz T. To differentiate or not--routes towards metastasis. Nat Rev Cancer 2012;12:425-36. [Crossref] [PubMed]
- Nieto MA, Huang RY, Jackson RA, et al. EMT: 2016. Cell 2016;166:21-45. [Crossref] [PubMed]
- Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol 2017;14:611-29. [Crossref] [PubMed]
- Nitulescu GM, Margina D, Juzenas P, et al. Akt inhibitors in cancer treatment: The long journey from drug discovery to clinical use Int J Oncol 2016;48:869-85. (Review). [Crossref] [PubMed]
- Vosgha H, Salajegheh A, Smith RA, et al. The important roles of miR-205 in normal physiology, cancers and as a potential therapeutic target. Curr Cancer Drug Targets 2014;14:621-37. [Crossref] [PubMed]
- Ye X, Brabletz T, Kang Y, et al. Upholding a role for EMT in breast cancer metastasis. Nature 2017;547:E1-3. [Crossref] [PubMed]
- Gomes BC, Rueff J, Rodrigues AS. MicroRNAs and Cancer Drug Resistance. Methods Mol Biol 2016;1395:137-62. [Crossref] [PubMed]
- Fischer KR, Durrans A, Lee S, et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 2015;527:472-6. [Crossref] [PubMed]
- Nikolova T, Kiweler N, Krämer OH. Interstrand Crosslink Repair as a Target for HDAC Inhibition. Trends Pharmacol Sci 2017;38:822-36. [Crossref] [PubMed]
- Korpal M, Kang Y. Targeting the transforming growth factor-beta signalling pathway in metastatic cancer. Eur J Cancer 2010;46:1232-40. [Crossref] [PubMed]


