TP53 mutations predict for poor survival in ALK rearrangement lung adenocarcinoma patients treated with crizotinib
Introduction
Lung cancer is one of the most common cancers and the leading cause of cancer death in the world (1). In the past decades, advances in molecular analysis and the development of targeted therapies have changed the identification of oncogenic drivers, the treatment and the survival for lung cancer. Anaplastic lymphoma kinase (ALK) rearrangement is one of the driver genes which detected in 3–7% of patients with non-small cell lung cancer (NSCLC) (2). For these patients, ALK tyrosine kinase inhibitors (TKIs) have been developed. Crizotinib is a first generation ALK-TKI that shows efficacy in the treatment of ALK rearrangement NSCLC (3). According to clinical studies of crizotinib (PROFILE 1005, PROFILE 1007, PROFILE1014), the major populations were lung adenocarcinoma (ADC) and an objective response rate (ORR) to crizotinib is approximately 60% and its median progression-free survival (PFS) is nearly 10 months (4). However, it is known that many cases ultimately acquired resistance to crizotinib (5). In addition, about 30% ALK-positive patients to the treatment of crizotinib demonstrate primary resistance which early progressive disease (PD) after the first month of treatment (6,7).
The mechanisms of acquired resistance included secondary mutations or copy number gain (CNG) in the ALK kinase domain and up regulation of bypass signaling pathways (8). However, to our knowledge, the mechanisms of primary resistance to ALK inhibitor for these patients remain elusive. And only some primary resistance mechanisms have been hypothesized, such as mutations of the EGFR pathway and BIM polymorphisms (9-11).
According to reports, the tumor suppressor of TP53 gene mutations occur in approximately 25–50% NSCLC patients (12,13). Ma et al. (14) has demonstrated regardless of EGFR and KRAS mutation status, non-disruptive TP53 mutations are independent markers of shorter overall survival (OS) in patients with advanced NSCLC. There is preclinical evidence in the human NSCLC cell lines NCI-H1299 and A549 showed a relationship between TP53 gene mutations and responsiveness to TKIs (15). Canale et al. (16) had reported that TP53 mutations reduce responsiveness to EGFR-TKIs and poor prognosis in EGFR-mutated NSCLC patients.
However, the relationship of TP53 gene status and the efficacy of crizotinib in ALK positive NSCLC patients was unclear. In this study we proposed to evaluate the role of TP53 mutation in ALK rearrangement advanced NSCLC patients that received crizotinib treatment. We analyzed the status of TP53 gene and its association with the effect of crizotinib in Chinese patients with ALK-positive NSCLC.
Methods
Patients
Eligible patients were required to have pathologically confirmed NSCLC and sufficient tissue for analysis. Clinical and pathologic data prospectively collected for analyses included age at diagnosis, gender, smoking status, and stage according to the new International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society multidisciplinary classification. This study was approved by the ethics committee of Fujian Provincial Cancer Hospital, Fujian Medical University Cancer Hospital, Fuzhou Fujian, China, and a written informed consent was obtained from each participant before the initiation of any study-related procedure.
Targeted next-generation sequencing
A total of 66 ALK positive patients treated with crizotinib. However, 28 specimens were not enough and 38 specimens were evaluated by NGS. For 38 patients, including parts of patients with ALK positive pts treatment before crizotinib, targeted region capture combined NGS was successfully performed in 21 patients. Genomic DNA sequencing libraries were prepared using the protocols recommended by the Illumina TruSeq DNA Library Preparation Kit. For samples close to the minimum input requirement, additional pre-capture PCR cycles were performed to generate sufficient PCR product for hybridization. The libraries were hybridized to custom-designed probes (Integrated DNA Technology) including all exons of 170 genes and selected intron of ALK, RET and ROS1 for the detection of Genomic rearrangements. DNA sequencing was performed on a HiSeq3000 sequencing system (Illumina, San Diego, CA) with 2×75 bp paired-end reads. The reads were aligned to the human genome build GRCh37 using BWA (a Burrows-Wheeler aligner). Somatic single nucleotide variant (sSNV) and indel calls were generated using MuTect and GATK, respectively. Somatic copy number alterations were identified with CONTRA. Genomic rearrangements were identified by the software developed in house analyzing chimeric read pairs.
Response evaluation
Patients received crizotinib treatment (250 mg, twice daily) and had clinical data available including general characteristics, treatment efficacy and adverse reactions to treatment. Imaging data were independently reviewed by authors to evaluate their treatment responses according to the Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1 PFS was calculated from the date of initiating targeted drugs treatment to radiologic or clinical observation of disease progression.
Statistical analyses
The response rate among subgroups and survival was described with Kaplan-Meier methodology and the log-rank test was used to compare survival among subgroups. Statistical analysis was performed using SPSS version 19.0 software (IBM, Armonk, NY, USA). All P values were 2-sided, and a P value of <0.05 was considered statistically significant.
Results
Patient characteristics
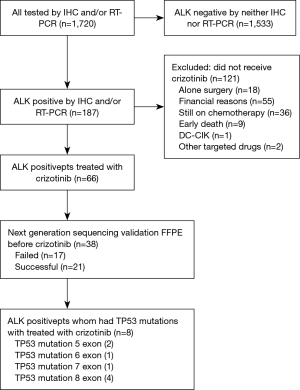
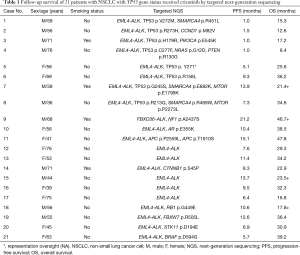
From July 2013 to May 2016, a total of 1,720 patients were enrolled in this study. Among them, 187 (10.87%) were identified as ALK-positive and 66 of them received oral crizotinib. Because of the tissue sample insufficient and specimen quality testing, 21 of them was successfully performed by targeted region capture combined NGS. The flow chart of the study design is shown in Figure 1. Clinic-pathological characteristics of patients were reported in Table 1. The majority of patients were male (52.38%, 11/21), less than 65 years old (61.90%, 13/21) and never smokers (80.95%, 17/21). All patients had a diagnosis of ADC (100%, 21/21). All patients received crizotinib (100%, 21/21). NGS detection found many accompanied genes, including mTOR pathway (PIK3CA p.E545K, FBXW7 p.R505L, STK11 p.D194E, PTEN p.R130G, MTOR p.E1799K, MTOR p.P2273L), RAS pathway (NRAS p.G12D), TP53 exon 5 (p.R158L, p.H179R), exon 6 (p.R213Q), exon 7 (p.G245S), exon 8 (p.Y271*, p.V272M, p.R273H, p.C277F) (Figure 2), SMARCA4 (p.R451L, p.E882K, p.R469W), AR (p.E355K), APC (p.P2559L, p.T1910S), CCND1 (p.M82V), CTNNB1 (p.S45P), BRAF (p.D594G), RB1 (p.G449E) and NF1 (p.A2437S).
Full table
TP53 mutation
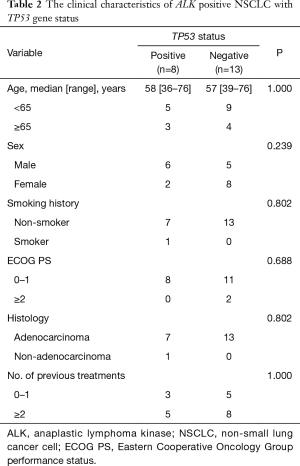
Out of the 21 patients with successfully targeted region capture combined NGS, 8 (38.10%) patients showed a TP53 mutation: 25.00% (2/8) were on exon 5, 12.50% (1/8) on exon 6, 12.50% (1/8) on exon 7, and 50.00% (4/8) on exon 8 (Table 1). The majority of patients were male (75.00%, 6/8), less than 65 years old (62.50%, 5/8) and never smokers (75.00%, 6/8). There were no significant differences noted with regard to age, sex, smoking history, ECOG PS, histology, number of previous treatments, between patients with and without the TP53 gene (Table 2).
Full table
TP53 mutation and response to crizotinib
ORR and disease control rate (DCR) for crizotinib in the entire case series were 61.90% and 71.43%, respectively. TP53 gene wild group was found to be significantly associated with a higher ORR and DCR to crizotinib with respect to TP53 gene mutation group. An ORR of 76.92% and a DCR of 84.62% were observed in patients with TP53 gene wild group, and an ORR of 37.50% and a DCR of 50.00% were observed in patients with TP53 gene mutation group (Table 3).

Full table
TP53 mutation and survival
Statistically significant difference was observed in terms of PFS and OS between TP53 gene wild group and mutation group patients (P=0.038, P=0.021, respectively). The PFS in TP53 gene mutation group patients was shorter than that in TP53 gene wild group (3.3 vs. 10.4 months). The OS of TP53 gene mutation group was poorer than TP53 gene wild group (21.4 vs. 34.2 months). PFS and OS of TP53 gene on exon 5 were 9.3, 36.2 months and 1.0, 17.2 months), respectively; PFS and OS of TP53 gene on exon 6 were 7.3 and 34.6 months; PFS and OS of TP53 gene on exon 7 were 13.9 months, more than 21.4 months; PFS and OS of TP53 gene on exon 8 were 5.1 and 25.6 months, 1.0 and 15.3 months, 1.5 and 12.6 months, and 1.0 and 8.4 months, respectively (Table 1, Figure 3).

Discussion
To our knowledge, our study is the first to explore the relationship between TP53 gene mutation status and the effect of crizotinib in Chinese patients with ALK-positive advanced NSCLC. NSCLC patients with ALK rearrangement are highly sensitive to crizotinib. Nowadays, a series of trial data (phase I–III) involving in ALK positive advanced NSCLC patients demonstrated the efficacy of crizotinib was good and adverse reactions could be tolerated (4,17,18). The data from East Asian patients also showed the ORR to crizotinib was 88% and its median PFS was 11.1 months (19). Unfortunately, some ALK-positive patients have been with shorter PFS after treatment with crizotinib which might be presence primary resistance to crizotinib. Up to now, the mechanisms of acquired resistance to ALK inhibitors can be divided into 2 types: ALK dominant or ALK non-dominant. ALK dominant includes secondary mutations and CNG in the ALK gene; ALK non-dominant includes the activation of bypass tracks, such as EGFR, MET, KIT, KRAS and IGF-1R (8). However, it is unclear that the causes of short PFS in ALK positive patients after crizotinib treatment. Some studies have explored the mechanisms of crizotinib primary resistance. Zhang et al. (9) reported patients with the BIM deletion polymorphism had a significantly shorter PFS (182 vs. 377 days, P=0.008) and lower ORR (44.4% vs. 81.7%, P=0.041) compared with those without in 69 ALK/ROS1 positive patients with crizotinib treatment. Doebele et al. (10) reported that a patient received re-biopsy after 61 days on crizotinib which showed stable disease. This biopsy demonstrated a lack of an ALK gene rearrangement by FISH, but the presence of an EGFR exon 21 mutation by direct sequencing. There is still a lack in the mechanisms reports of primary resistance to crizotinib with ALK-positive NSCLC.
It is reported that TP53 gene mutations occur in approximately 50% of lung cancer patients and are more common in squamous cell lung carcinoma than in lung ADC (12). In the lung ADC cancer, TP53 mutation rates ranges from 25% to 40% (20,21). In addition, some reports showed the prevalence of ALK-rearranged with p53 mutation was about 9.1% (1/11) to 28.6% (2/7) (22,23). In our study, we observed a mutation percentage of 38.1% in 21 ALK positive ADC patients. Therefore, coexisting with TP53 mutations in ALK positive ADC patients is still further to explore in a larger research, especially for comparing ALK rearrangement patients with ALK negative patients. The p53 protein regulates cellular response to a variety of cellular stress signals by inducing cell cycle arrest (24). The normal function disruption of p53 disrupts this cellular response, leading to possible malignant cell transformation. Gene mutations lead to a loss of p53 functions, however non-disruptive mutations could remain some of the p53 protein functional properties (25). Therefore, we analyzed the relationship of efficacy and survival between ALK positive and TP53 gene status with crizotinib therapy.
It is the first time to put forward that patients with TP53 gene mutation can no response to crizotinib. In our study, TP53 gene mutation group was found to be significantly associated with a lower ORR (76.92% vs. 37.50%) and DCR (50.00% vs. 84.62%) to crizotinib with respect to TP53 gene wild group. In addition, significant shorter PFS (3.3 vs. 10.4 months, P=0.038) and OS (21.4 vs. 34.2 months, P=0.021) were observed in patients with TP53 gene mutation group patients than TP53 gene mutation group. In particular, 4 of 8 TP53 mutated patients showed PD within two months of crizotinib therapy. Therefore, we think ALK positive patients with TP53 gene mutation would influence the efficacy of crizotinib treatment. Likewise, in EGFR mutation populations, Canale et al. (16) also demonstrated TP53 mutations were associated with a significantly lower DCR respect to wild patients. However, no statistically significant difference was observed in terms of PFS and OS between two groups. Their most significant result was that TP53 exon 8 mutation was associated with the worst prognosis. Our study also found ALK positive patients with TP53 exon 8 mutation were poorer survival than other mutation types. The TP53 mutant type has been found to be an important factor for gefitinib-induced apoptosis in NSCLC cell lines and reduces gefitinib-induced apoptosis (26). We think that TP53 could have a role as prognostic factor to ALK inhibitors, rather than predictive factor. In the future, we should carry out well-designed prospective multicenter studies to demonstrate the correlation between TP53 gene status and ALK inhibitors including other generations in patients with ALK-positive disease.
We also recognize that there are several limitations to our study. First, due to a small sample, the conclusions should be bias a certain extent. Second, this was a retrospective study, and therefore selection bias was inevitable. Hence, future research will continue to increase the population in the hope of reducing the bias to some extent.
In conclusion, we described the relationship between TP53 gene and the efficacy of crizotinib in ALK rearrangement patients. And the TP53 mutant patients were associated with poor ORR and shorter PFS. However, more work and further studies should be performed to explore the significance of TP53 gene in patients with ALK-positive NSCLC who were treated with ALK inhibitors.
Acknowledgements
Funding: This study was supported in part by grants from the National Clinical Key Specialty Construction Program (2013); Leading Project Foundation of Science Department of Fujian Province (2016Y0019); Leading Project Foundation of Science Department of Fujian Province (2015Y0011).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: This study was approved by the ethics committee of Fujian Provincial Cancer Hospital, Fujian Medical University Cancer Hospital, Fuzhou Fujian, China, and a written informed consent was obtained from each participant before the initiation of any study-related procedure.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Ettinger DS, Wood D, Akerley W, et al. Non-Small Cell Lung Cancer, Version 6. 2015. J Natl Compr Canc Netw 2015;13:515-24. [Crossref] [PubMed]
- Sullivan I, Planchard D. Treatment modalities for advanced ALK-rearranged non-small-cell lung cancer. Future Oncol 2016;12:945-61. [Crossref] [PubMed]
- Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013;368:2385-94. [Crossref] [PubMed]
- Toyokawa G, Seto T. Updated Evidence on the Mechanisms of Resistance to ALK Inhibitors and Strategies to Overcome Such Resistance: Clinical and Preclinical Data. Oncol Res Treat 2015;38:291-8. [Crossref] [PubMed]
- Gainor JF, Dardaei L, Yoda S, et al. Molecular mechanisms of resistance to first- and second-generation ALK inhibitors in ALK-rearranged lung cancer. Cancer Discov 2016;6:1118-33. [Crossref] [PubMed]
- Steuer CE, Ramalingam SS. ALK-positive non-small cell lung cancer: mechanisms of resistance and emerging treatment options. Cancer 2014;120:2392-402. [Crossref] [PubMed]
- Katayama R, Shaw AT, Khan TM, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung cancers. Sci Transl Med 2012;4:120ra17. [Crossref] [PubMed]
- Zhang L, Jiang T, Li X, et al. Clinical features of Bim deletion polymorphism and its relation with crizotinib primary resistance in Chinese patients with ALK/ROS1 fusion-positive non-small cell lung cancer. Cancer 2017. [Epub ahead of print]. [PubMed]
- Doebele RC, Pilling A, Aisner D, et al. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clin Cancer Res 2012;18:1472-82. [Crossref] [PubMed]
- Wang W, Jiang X, Song Z, et al. Patients harboring EGFR mutation after primary resistance to crizotinib and response to EGFR-tyrosine kinase inhibitor. Onco Targets Ther 2016;9:211-5. [Crossref] [PubMed]
- Mogi A, Kuwano H. TP53 mutations in non small cell lung cancer. J Biomed Biotechnol 2011;2011:583929. [Crossref] [PubMed]
- Deben C, Deschoolmeester V, Lardon F, et al. TP53 and MDM2 genetic alterations in non-small cell lung cancer: Evaluating their prognostic and predictive value. Crit Rev Oncol Hematol 2016;99:63-73. [Crossref] [PubMed]
- Molina-Vila MA, Bertran-Alamillo J, Gasco A, et al. Nondisruptive p53 mutations are associated with shorter survival in patients with advanced non-small cell lung cancer. Clin Cancer Res. 2014;20:4647-59. [Crossref] [PubMed]
- Rho JK, Choi YJ, Ryoo BY, et al. p53 enhances gefitinib-induced growth inhibition and apoptosis by regulation of fas in non-small cell lung cancer. Cancer Res 2007;67:1163-9. [Crossref] [PubMed]
- Canale M, Petracci E, Delmonte A, et al. Impact of TP53 Mutations on Outcome in EGFR-Mutated Patients Treated with First-Line Tyrosine Kinase Inhibitors. Clin Cancer Res 2017;23:2195-202. [Crossref] [PubMed]
- Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010;363:1693-703. [Crossref] [PubMed]
- Solomon BJ, Mok T, Kim DW, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med 2014;371:2167-77. [Crossref] [PubMed]
- Lu S, Mok T, Lu Y, et al. Phase 3 study of first-line crizotinib vs pemetrexed cisplatin/carboplatin (PCC) in East Asian patients (pts) with ALK+ advanced non-squamous non-small cell lung cancer (NSCLC). J Clin Oncol 2016;34:abstr 9058.
- Ma X, Le Teuff G, Lacas B, et al. Prognostic and predictive effect of TP53 mutations in non-small cell lung cancer patients from adjuvant cisplatin-based therapy randomized trials: A LACE-bio pooled analysis. J Thorac Oncol 2016;11:850-61. [Crossref] [PubMed]
- Halvorsen AR, Silwal-Pandit L, Meza-Zepeda L A, et al. TP53 mutation spectrum in smokers and never smoking lung cancer patients. Front Genet 2016;7:85. [Crossref] [PubMed]
- Tuononen K, Kero M, Mäki-Nevala S, et al. ALK fusion and its association with other driver gene mutations in Finnish non-small cell lung cancer patients. Genes Chromosomes Cancer 2014;53:895-901. [Crossref] [PubMed]
- Inamura K, Takeuchi K, Togashi Y, et al. EML4-ALK lung cancers are characterized by rare other mutations, a TTF-1 cell lineage, an acinar histology, and young onset. Mod Pathol 2009;22:508-15. [Crossref] [PubMed]
- Zilfou JT, Lowe SW. Tumor suppressive functions of p53. Cold Spring Harb Perspect Biol 2009;1:a001883. [Crossref] [PubMed]
- Poeta ML, Manola J, Goldwasser MA, et al. TP53 mutations and survival in squamous-cell carcinoma of the head and neck. N Engl J Med 2007;357:2552-61. [Crossref] [PubMed]
- Deben C, Van den Bossche J, Van Der Steen N, et al. Deep sequencing of the TP53 gene reveals a potential risk allele for non-small cell lung cancer and supports the negative prognostic value of TP53 variants. Tumour Biol 2017;39:1010428317694327. [Crossref] [PubMed]