Next-generation sequencing for identifying genetic mutations in adults with bronchiectasis
Introduction
Bronchiectasis is characterized by irreversible bronchial dilatation resulting from airway infection and inflammation (1). Various etiologies have been attributable to bronchiectasis (2-4), but whether genetic mutations are underlying causes is unclear. Certain mutations (e.g., RSPH1, RSPH4A, RSPH9 and DNAI1) related to primary ciliary dyskinesia (PCD) (5-8), immunodeficiency (9), and cystic fibrosis (CF) (10,11) have been implicated in the pathogenesis of bronchiectasis. However, previous studies were limited by relatively small sample sizes (7,9-11), selective mutation analysis (5-9,11) and children-predominant population (5,6), which limited the generalizability of findings (12).
Next-generation sequencing (NGS) with Ion Torrent PGMTM platform (Life Technologies Inc., USA) has fueled the testing of various diseases (10). Compared with Sanger sequencing, NGS is simpler and convenient, has higher detection accuracy and faster responses at lower cost. NGS confers a greater likelihood for identifying novel mutations (13-16), which might unravel how mutations are implicated in bronchiectasis.
Airway epithelium plays pivotal roles in maintaining host-defense to pathogen invasion. Ciliary defects associated with PCD and cystic fibrosis transmembrane conductance regulator (CFTR) disorders have been linked to bronchiectasis pathogenesis. Defects of RSPH1, RSPH9 (17) or DNAH11 genes (18) could have resulted in bronchiectasis due to impaired ciliary ultrastructure and/or beating.
In view that pathogenic mutations associated with defective airway host-defense predispose to bronchiectasis, we compared mutation frequency between bronchiectasis patients and healthy subjects in Chinese Han ethnicity, identified pathogenic mutations, and determined the clinical characteristics of bronchiectasis with biallelic mutations.
Methods
Subjects
Adults with bronchiectasis, diagnosed with chest high-resolution computed tomography (HRCT), were recruited from The First Affiliated Hospital of Guangzhou Medical University between September 2012 and 2015. Cystic fibrosis was not routinely screened because of extremely low prevalence in China. Healthy subjects with normal chest X-ray and no prior history or symptoms of chronic respiratory diseases (including asthma, chronic obstructive pulmonary disease, etc.) whose data were anonymized before blood collection were enrolled from our health check-up center. None of the subjects had malignancy, severe systemic disorders (e.g., stroke), or upper airway infection or antibiotic use within 4 weeks. Ethics approval was obtained from Ethics Committee of The First Affiliated Hospital of Guangzhou Medical University. Written informed consent was obtained.
Clinical assessments
Venous blood was withdrawn, followed by gradient centrifugation in heparinized tubes. Genomic DNA was extracted using commercialized kits (Qiagen, Valencia, CA).
For bronchiectasis patients, clinical investigations included etiology (4), chest HRCT, spirometry (19) and sputum bacteriology (20). Chest HRCT was assessed using the modified Reiff score (21). Fresh sputum was immediately sent for microbiology assessment. We calculated Bronchiectasis Severity Index (BSI) to determine disease severity (22) (See online Supplement text).
Mutation screening
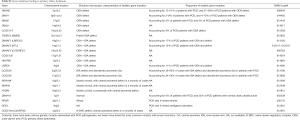
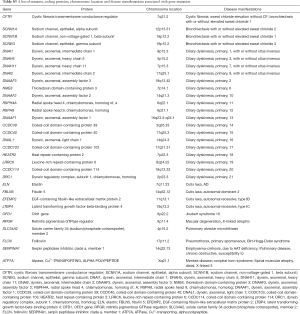
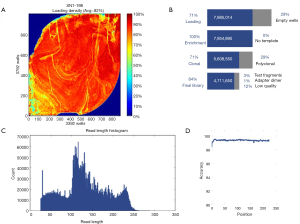
Mutations of multiple genes have been described as a cause of bronchiectasis (23-25). Thirty-two clinically important candidate genes of cystic fibrosis (CFTR, SCNN1A, SCNN1B and SCNN1G), PCD (CCDC103, CCDC114, CCDC164, CCDC39, CCDC40, DNAAF1, DNAAF2, DNAAF3, DNAH11, DNAH5, DNAI1, DNAI2, DNAL1, HEATR2, LRRC6, OFD1, RPGR, RSPH4A, RSPH9, and NME8) and other lung diseases (EFEMP2, ELN, FBLN5, FLCN, LTBP4, SERPINA1, ATP7A and SLC34A2) were screened in our study (see the detailed panel in Tables S1-S3 in Online Supplement text). Data analysis was performed with the hypothesis that bronchiectasis might be a single-gene disorder. The conservation and pathogenicity of the mutations were also determined (Figure 1).

Full table
Full table
Full table
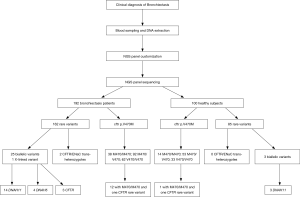
In this study, sequencing libraries were constructed using 10 ng of the genomic deoxyribonucleic acid (DNA). The DNA was amplified using a custom-designed Ion AmpliSeq™ Panel (32 genes) (Life Technologies, Grand Island, NY, USA) and barcoded by using the Ion Xpress Fragment Library Kit and Ion Xpress™ Barcode Adapters (Life Technologies, USA). The DNA libraries were then amplified with the OneTouch Template 200 Kit on the Ion One Touch™ 2 (Life Technologies, USA) and enriched on the Ion OneTouch™ ES. The enriched libraries were sequenced on an Ion Torrent PGM platform (Life Technologies Inc., USA) after annealing the sequencing primer, binding with the Ion PGM™ Sequencing Polymerase and loading on the Ion 318™ v2 Chip according to the manufacturer’s protocols. Software including Torrent Suite™ 4.0 (Life Technologies Inc., USA) and wANNOVAR (web ANNOVAR) were applied to perform bioinformatic analysis to annotate the genetic variants in terms of disease phenotypes. Finally, all likely pathogenic variants were confirmed by Sanger sequencing.
Statistical analysis
Statistical analysis was performed using SPSS 16.0 (SPSS Inc., Ill, USA). We presented numerical data as mean ± standard deviation or median (interquartile range), and categorical data as number (percentage). Independent t-test or Mann-Whitney test was employed for two-group comparisons. Chi-square test was performed to compare rates for two groups. P<0.05 was deemed statistically significant for all analyses unless otherwise stated.
Results
Patient characteristics
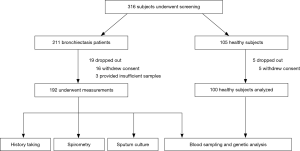
Of 316 subjects screened, 24 withdrew consent. Data from 192 bronchiectasis patients and 100 healthy subjects were analyzed (Figure 2).
Our cohort consisted of mainly middle-aged adults with a female predominance. Twenty-two (11.5%) and 11 patients (5.7%) reported a family history of chronic respiratory diseases and bronchiectasis, respectively. The dominant etiology was idiopathic (31.8%), followed by post-infectious (27.6%) (Table 1).

Full table
Panel performance of candidate genes
Sequencing of 32 genes (see Table S3 in Online Supplement text) yielded a mean depth of ~200× and coverage of 99.3% (Figure S1). Exons with low (<20-fold) or no coverage were subject to Sanger sequencing to achieve 100% coverage. Pathogenic and rare mutations of unknown significance were confirmed with Sanger sequencing.
We identified 162 and 85 rare variants in bronchiectasis patients and healthy subjects, respectively. In bronchiectasis, 25 (15.4%), 117 (72.2%) and 18 (11.1%) rare variants were associated with CFTR/epithelial sodium channel (ENaC), PCD and other relevant genes, respectively (see Tables S4,S5 in Online Supplement text).

Full table

Full table
CFTR and ENaC mutations
We examined CFTR, SCNN1A, SCNN1B and SCNN1G mutations via screening the published pathogenic mutations from databases, followed by identification with single nucleotide polymorphism frequency <0.1% and other novel mutations. Pathogenicity was predicted with the software Sift (http://sift.jcvi.org) and Polyphen2 (http://genetics.bwh.harvard.edu/pph2/).
Overall, 49 bronchiectasis patients (25.5%) and 26 healthy subjects (26.0%) harbored at least one rare CFTR variant, respectively (see Tables S4,S5 in Online Supplement text). Forty-three bronchiectasis patients (22.4%) harbored 15 CFTR variants, whereas 6 patients (3.1%) harbored six SCNN1A and SCNN1B variants. Twenty-five healthy subjects (25.0%) harbored 13 CFTR variants, whereas 1 (1.0%) harbored SCNN1B variant. We identified five novel variants: c.1252C>A(p.P418T), c.205G>A(p.D69N) and c.953T>G(p.I318S) for SCNN1A, and c.3592G>A(p.V1198M) and c.4262T>A(p.V1421E) for CFTR.
Eighteen bronchiectasis patients (9.4%) harbored c.1666A>G(p.I556V) in CFTR variants (exon 12). No significant differences in c.1666A>G frequency were found compared with healthy subjects (9.0%, P=0.920). An identical stop-gain CFTR mutation, c.1909C>T(p.Q637X), was identified in two bronchiectasis patients but none of the healthy subjects. SCNN1A and SCNN1B variants were identified in 6 bronchiectasis patients (3.1%) and 1 healthy subject (1.0%). The frequency of heterozygous mutations was comparable between healthy subjects and bronchiectasis patients (P=0.740) (Table S6).
Full table
Notably, four bronchiectasis patients harbored biallelic CFTR variants. Patient B168 harbored c.91C>T(p.R31C) and c.2684G>A(p.S895N) variants, patient B077 harbored IVS8 5T and c.1666A>G(p.I556V) variants, patient B179 harbored homozygous IVS8 5T variants, and patient B180 harbored c.1666A>G(p.I556V) and c.3289C>T(p.R1097) variants (Table 2). However, none of the healthy subjects harbored biallelic CFTR variants.
Full table
Moreover, patient B142 harbored a c.T953G(p.I318S) missense mutation of SCCN1A and a c.A1666G(p.I556V) mutation of CFTR. Patient B180 harbored c.A1666G(p.I556V) and c.C3289T(p.R1097C) mutations of CFTR, and c.A806T(p.Y269F) of SCCN1B. None of the healthy subjects harbored CFTR/ENaC trans-heterozygote mutations.
p.V470M polymorphism
We identified 130 bronchiectasis patients (67.7%) who harbored p.M470 variants of CFTR. Homozygous p.M470 variants were identified in 38 bronchiectasis patients (29.2%), whereas heterozygous p.M470 variants in 92 bronchiectasis patients (70.7%). Sixty-seven healthy subjects (67.0%) harbored p.M470 variants. Of these, 14 subjects (20.8%) harbored homozygous p.M470 and 53 patients (79.1%) harbored heterozygous variants. No significant difference in the number of bronchiectasis patients with p.M470 carriers (P=0.902) or homozygous variants was found (P=0.209).
Of 92 bronchiectasis patients with heterozygous p.M470 variants, 20 (21.7%) harbored other rare CFTR variants. Of 38 bronchiectasis patients with homozygous p.M470 variants, 12 (31.5%) harbored miscellaneous CFTR mutations. Miscellaneous CFTR mutations were detected in 12 (22.6%) of 53 healthy subjects with heterozygous p.M470 variants and 1 (7%) of 14 healthy subjects with homozygous p.M470 variant.
Homozygous p.M470 and other CFTR variants were more frequently observed in bronchiectasis patients than in healthy subjects (6.3% vs. 1.0%, P=0.039) (Table 3).

Full table
PCD gene mutations
We detected 117 rare variants of PCD-associated genes in 101 bronchiectasis patients. When analyzing 20 PCD-associated genes, we identified 36 DNAH11 variants (18 novel variants), 22 DNAH5 variants (11 novel variants), nine CCDC40 variants, seven HEATR2 variants, six variants each of DNAAF2 and DRC1, four variants each of RSPH9 and NME8, three variants each of CCDC114, CCDC39, DNAAF1, DNAAF3, LRRC6, RPGR and DNAI2, two variants of DNAI1, and one CCDC103 variant. Fifty-nine variants (27.3%) were identified for DNAH11 and DNAH5 (Table S2). Moreover, we found eight carriers of c.G1394A(p.R465Q) variant (rs201303241) for HEATR2 in exon 6, which was absent in healthy subjects. Five healthy subjects harbored c.G1270A(p.A424T) variants in exon 6 which were not identified among bronchiectasis patients.
Twenty bronchiectasis patients (17.1%) harbored biallelic variants, of whom 14 (70.0%) had biallelic DNAH11 variants and four (16.0%) harbored biallelic DNAH5 variants (Table 3). Patients B39 and B176 harbored homozygous variants of c.1394G>A(p.R465Q) for HEATR2 and c.13373C>T(p.P4458L) for DNAH11, respectively. Patient B188 harbored c.518G>A(p.G173D) for RPGR. All these bronchiectasis patients had dual missense mutations, except for a single missense mutation plus splice mutation in patient B002, and non-sense mutation in patient B012.
Sixty-nine variants of PCD-associated genes were detected in 60 healthy subjects. When analyzing 15 PCD-associated genes, we identified 36 DNAH11 variants, 16 DNAH5 variants, 11 LRRC6 variants, six variants each for CCDC40, DRC1, and HEATR2, five variants each for DNAI2 and RPGR, four DNAAF3 variants, three variants each for DNAAF1, DNAAF2 and NME8, and two variants each for CCDC114, CCDC39 and RSPH9. Of these, 52 variants (45.6%) were identified for DNAH11 and DNAH5.
Of 60 healthy subjects with missense mutations, 3 (5%) harbored biallelic DNAH11 variants (Table 3).
Clinical manifestations of patients with dual mutations
Twenty-five bronchiectasis patients (13.0%) and three healthy subjects (3.0%) harbored biallelic rare variants of an identical gene (χ2=7.62, P=0.006). Of 25 bronchiectasis patients, 4 (16.0%) harbored biallelic CFTR variants, 4 (16.0%) harbored biallelic DNAH5 variants and 14 (56.0%) harbored biallelic DNAH11 variants (Table 4).

Full table
Of 14 patients with biallelic DNAH11 variants, 3 (21.4%) had severe bronchiectasis. The most common etiology was idiopathic (57.1%), followed by post-tuberculous (21.4%). Mean HRCT score was 8.3. Mean forced expiratory volume in one second (FEV1) was 77.3% predicted. Eleven (78.6%) patients had symptom onset for 20–50 years. Eight, six and two patients had cystic bronchiectasis, Pseudomonas aeruginosa isolated from sputum, and a family history of bronchiectasis, respectively.
Of four patients with biallelic DNAH5 variants, 3 (75.0%) had severe bronchiectasis. The most common etiology was idiopathic (75.0%). The mean number of bronchiectatic lobes was 3.8 and HRCT score 8.0. Mean FEV1 was 71.7% predicted. Three patients (75.0%) had symptom onset for 20–50 years. Three, one and two patients had cystic bronchiectasis, Pseudomonas aeruginosa isolated from sputum, and a family history of bronchiectasis, respectively.
Of four patients with biallelic CFTR variants, 1 (20.0%) had severe bronchiectasis, 3 (60.0%) had idiopathic bronchiectasis. Mean HRCT score was 6.2. Four patients had cystic bronchiectasis. Pseudomonas aeruginosa was isolated in one case. No family history of bronchiectasis was documented.
Patient B180 who harbored biallelic CFTR variants and one ENaC variant had greater disease severity (BSI =10) than patient B140, the carrier of CFTR plus ENaC variant (BSI =2). As a carrier of SCNN1B, Patient B180 had more severe bronchiectasis compared with patient B077 (BSI =2) and B168 (BSI =5), carriers of biallelic CFTR variants without ENaC mutation.
Bronchiectasis etiology in patients with biallelic mutations
Of 192 bronchiectasis patients, 26 (13.4%) harbored biallelic mutations or an X-linked pathogenic mutation. The underlying causes of bronchiectasis were idiopathic, post-infectious, gastroesophageal reflux, immunodeficiency and asthma in 16, 6, 2, 1 and 1 each of bronchiectasis patients, respectively.
Discussion
Our study analyzed 32 genes associated with CF and PCD in bronchiectasis. Patients harboring biallelic rare mutations accounted for 13.4% of the cohort. Compared with healthy subjects, homozygous mutation of p.V470M or ENaC plus other CFTR mutations were more frequently observed in patients with bronchiectasis. Compared with bronchiectasis patients with biallelic DNAH11 or CFTR mutations, those with biallelic DNAH5 mutation seemed more likely to have greater bronchiectasis severity.
Altered mucus property arising from CFTR defects and/or ENaC over-expression has been linked to bronchiectasis. Fajac et al. (26) identified five amino-acid changes (p.Ser82Cys, p.Asn288Ser and p.Pro369Thr in ENaCβ; p.Gly183Ser and p.Glu197Lys in ENaCγ) in eight bronchiectasis patients with heterozygous ENaC mutations, and one amino-acid change (p.Ser82Cys) in one patient with CFTR mutation. In our study, none of the biallelic CFTR variants was identified in healthy subjects; three of six variants (50.0%) that were identified among bronchiectasis patients were novel ENaC variants (including SCNN1A, SCNN1B and SCNN1G), whereas SCNN1B variant was detected in healthy subjects only (Table S4). However, SCNN1A mutations alone might be insufficient to result in bronchiectasis, because only the p.W493R-SCNN1A and p.F61L-SCNN1A mutations predisposed to CF-like lung disease via hyperreactive sodium absorption (27). Gain-of-function (auto-dominant) mutation of ENaC predisposed to CF via increased sodium absorption, whereas loss-of-function (auto-recessive) mutation did not invariably result in airway diseases despite perturbed ion transportation (28,29). Nonetheless, whether ENaC mutations belonged to gain- or loss-of-function mutation awaits further investigation.
One of the major pulmonary manifestations of cystic fibrosis is bronchiectasis. The frequency of cystic fibrosis is approximately 1 in 3,500 newborn Caucasians. However, cystic fibrosis has rarely been reported in Chinese patients, and there has been sparse research evidence from China. To our knowledge, fewer than 30 patients have been reported in the Chinese population. In our study, we identified four patients with biallelic CFTR variant but none of the biallelic CFTR variant was identified in healthy subjects, suggesting that CFTR mutations may have played a role in the pathogenesis of bronchiectasis in China. It has been reported that M470 CFTR proteins have a 1.7-fold increased intrinsic chloride channel activity compared with V470 CFTR proteins (30). Additionally, 90% of bronchiectasis patients with mutations harbored the more functional M470 allele (31). In our study, 12 bronchiectasis patients simultaneously harbored M470/M470 and other CFTR variants, as did in one healthy subject. This indicated that homozygous M470 mutation plus other CFTR variants might more readily predispose to bronchiectasis, although further investigations for validation are merited.
ENaC and CFTR mutations reportedly conferred synergistic effects on the development of diffuse bronchiectasis (32). Patient B142 harbored missense mutation c.T953G(p.I318S) of SCCN1A and c.A1666G(p.I556V) of CFTR; patient B180 harbored c.A1666G(p.I556V) and c.C3289T(p.R1097C) mutation of CFTR, and c.A806T(p.Y269F) of SCCN1B. None of these variants was detected among healthy subjects. Moreover, patient B180 who harbored biallelic CFTR mutations plus SCNN1A variant had greater disease severity (BSI =10) than patient B140 (BSI =2) who had CFTR plus SCNN1B variant, and patient B077 (BSI =2) and B168 (BSI =5) who harbored dual CFTR variants but no ENaC variant.
Several variants of PCD-associated genes leading to abnormal ciliary motility have been previously reported. DNAH5 mutation accounted for 27–38% of PCD patients with outer dynein arm defects (33). Despite a smaller proportion of PCD cases being affected and more subtle changes in ciliary ultrastructure, DNAH11 mutation contributed to rigid and hypermotile cilia which hampered mucus clearance (18). Other mutations, including RSPH9 and DNAI1, have been implicated in the pathogenesis of bronchiectasis (17). Through sequencing of PCD-associated auto-recessive mutations (apart from OFD1 and RPGR located in X-chromosomes), we identified 10.9% (n=21) of patients with at least one mutation (DNAH11: 7.3%, DNAH5: 2.6%). Of these, 14 cases harbored dual biallelic DNAH11 heterozygous mutation and four harbored dual biallelic DNAH5 heterozygous mutation (Table 2). Nonetheless, none of the bronchiectasis patients harbored DNAI1 mutation. Intriguingly, patients with DNAH5 mutation had greater disease severity compared with those with DNAH11 mutation (mean BSI: 8.8 vs. 4.2), which corroborated with the findings that DNAH5 mutation led to outer dynein arm defects and that DNAH11 mutation may have limited negative impacts on ciliary ultrastructure (normal or near-normal ultrastructural changes) or motility. Moreover, we have identified eight bronchiectasis patients with HEATR2 mutation (c.1394G>A (p.R465Q), heterogeneous mutation in seven cases), which was absent in healthy subjects, indicating that screening for HEATR2 mutation should be recommended.
Bronchiectasis is a heterogeneous disease with limited therapeutic options (3). Despite exhaustive etiologic assessment, underlying causes cannot be determined in approximately 50% of cases (34). Identifying etiologies might lead to changes in patient’s management (35). In our study, genetically susceptibility was revealed in 13.4% of patients with idiopathic bronchiectasis. Therefore, genetic testing complements conventional etiologic assessment by offering novel molecular information which would stimulate more in-depth research regarding host-defense defects in, and provide novel targets for future interventions of, bronchiectasis.
Nevertheless, we have included uniformly the Chinese Han population, which have limited the generalizability of our findings, as evidenced by the predominance of CFTR mutation in both healthy subjects and bronchiectasis patients. We might have underestimated some clinically significant mutations. Our findings cannot address whether other pathways (e.g., immunodeficiency, airway inflammation) are associated with bronchiectasis. Exploration of the association between genetic mutations of the immune system may help elucidate the mechanisms underlying how immunodeficiency contributes to bronchiectasis pathogenesis. The consequence of mutations has yet to be verified with sweat tests, nasal potential difference measurement and ciliary tests. The putative links between genetic mutations and bronchiectasis await verification with in vivo and in vitro models.
In conclusion, we have identified mutations associated with defective airway host-defense in bronchiectasis. Further development and functional validation of genetic testing for bronchiectasis is warranted.
Methods
Summarized methods for determining bronchiectasis etiologies
Patients were meticulously inquired the past and present history, followed by physical examination, chest (and nasal, if indicated) HRCT assessment, spirometry, diffusing capacity measurement, venous blood sampling (blood routine test and immunoglobulins), sputum collection and culture, saccharine test and simplified reflux questionnaire interview. Patients susceptible of having gastroesophageal reflux disease (GERD), aspergillosis, asthma or chronic obstructive pulmonary disease (COPD) underwent 24-hour esophageal PH monitoring, serum total and Aspergillus fumigatus-specific IgE test, and bronchial dilation test, respectively. Hemagglutinin and auto-antibodies were tested for susceptible cases with diffuse panbronchiolitis and connective tissue diseases, respectively. Other diagnostic tests (e.g., bronchoscopy, gastroscopy) could be performed if appropriate (1,4,36).
Sputum bacteriology
Fresh sputum was sampled during hospital visits. Following removal of debris in oral cavity, patients expectorated into sterile container for bacterial culture. Hypertonic saline (3–5%) induction, previously validated, was applied as appropriate. Sputum was sent for bacterial culture within 2 hours of sampling.
Blood and chocolate agar plates (Biomeurix, France) were adopted as culture media. Fresh sputum was homogenized with SPUTASOL (Oxoid SR089A, Cambridge, UK) and serially diluted with natural saline at concentrations of 10−4, 10−5 and 10−6. This was followed by addition of 10µl respective diluent to the plates with micropipette tube and inoculation using 10 µL standardized rings. Plates were positioned in thermostatic box containing 5% carbon dioxide at 37 degrees for overnight incubation (20).
Assessment of chest HRCT scores
Chest HRCT at collimation of 2 mm or less within 12 months was captured. Bronchiectasis was diagnosed if the internal diameter of bronchi was greater than accompanying pulmonary artery. Miscellaneous signs of bronchiectasis included the lack of normal bronchial tapering along travel on sequential slices and (or) visible bronchi within 10 mm to the pleura. The HRCT score was assessed on a lobar basis, with lingular lobe being regarded as a separate lobe. For an individual lung lobe, the radiological severity of bronchiectasis was scaled by using modified Reiff score. The maximal total score was 18 for a total of six lobes (1,4,36).
Spirometry
Spirometry was conducted by using spirometers (QUARK PFT, COSMED Co. Ltd., Milan, Italy) according to American Thoracic Society/European Respiratory Society guidelines. Variation between the best two maneuvers was <5% or 200 mL in FVC and FEV1, with maximal values being reported. Predicted values were derived from the model by Guan et al. (19,37).
Bronchiectasis severity index
Bronchiectasis severity index (BSI), a novel tool previously validated, was applied to determine the severity of bronchiectasis. The BSI was a composite of clinical parameters, including the age, body-mass index, prior exacerbation and prior hospitalization in the preceding year, modified Medical Research Council dyspnea score, FEV1 predicted%, Pseudomonas aeruginosa infection, colonization with other potentially pathogenic microorganisms (PPMs, including Haemophilus influenzae, Haemophilus parainfluenzae, Staphylococcus aureus, Klebsiella spp and other clinically significant bacteria) and the number of bronchiectatic lobes. The BSI of ≤4, 5–8 and ≥9 denoted mild, moderate and severe bronchiectasis, respectively (22).
Strategies for screening mutants associated with bronchiectasis
Candidate mutants related to bronchiectasis pathogenesis were screened from literature reports (23-25) and official websites, including Human Gene Mutation Database (http://www.hgmd.cf.ac.uk/ac/index.php) and Online Mendelian Inheritance in Man (http://omim.org/). The conservation and pathogenicity of mutants were subsequently determined (Figure S1).
First, we performed a pilot analysis on the sequencing data following gene sequencing with the Ion TorrentTM (Life Technologies Inc., NY, USA), which provided the automatic analyzing systems, the variantCallerTM and coverageAnalysisTM (Life Technologies Inc., NY, USA). The summary of mutation would be displayed through computerized automatic comparisons. We employed the wANNOVAR website for annotating the functional consequences of gene mutants from the sequencing data. This has allowed researchers to screen for candidate mutants using the databases as delineated previously.
Next, the primary source file (*.bam) was generated and subsequently transformed into a readable Excel file (*.xls) following annotation, which was applied for the detailed comparison on candidate mutants with the original database to minimize errors (online: http://jtd.amegroups.com/public/system/jtd/supp-jtd.2018.04.134-1.pdf; http://jtd.amegroups.com/public/system/jtd/supp-jtd.2018.04.134-2.pdf). This would allow investigators to exclude the false-positive mutant loci with poor sequencing quality.
Finally, we targeted at screening for clinically significant mutations including missense, nonsense, frameshift mutation which were likely to lead to conformational or functional changes in proteins. Factors such as the potential pathogenicity (from previously published literature and databases such as NCBI and HGMD), the frequency of SNP (using the 1000 Genome database) and conversation (using SIFT and Polyphen2 software) should be taken into account when determining the panel of candidate mutant genes. In case no significant mutations were identified, we would then proceed to screen for candidate mutants according to their introns or UTR, as well as candidate mutants in the loci with synonymous mutation or splice mutation.
The panel of 32 candidate genes consisted of genes associated with bronchiectasis (CFTR, SCNN1A, SCNN1B and SCNN1G), PCD (CCDC103, CCDC114, CCDC164, CCDC39, CCDC40, DNAAF1, DNAAF2, DNAAF3, DNAH11, DNAH5, DNAI1, DNAI2, DNAL1, HEATR2, LRRC6, OFD1, RPGR, RSPH4A, RSPH9, and TXNDC3) and cystic fibrosis (CF) pathogenesis (EFEMP2, ELN, FBLN5, FLCN, LTBP4, SERPINA1, and ATP7A), and other pathogenic genes (PAM and SLC34A2) (Tables S1-S3).
Deoxyribonucleic acid extraction and mutation identification
Proband deoxyribonucleic acid (DNA), extracted from heparinized venous blood, was detected with NGS platform using customized Ion AmpliSeqTM Panel (Life Technologies Inc., NY, USA). This panel was designed to identify disease-causing mutations in exons and exon-intron boundaries (±50 bp) of bronchiectasis genes and those associated with PCD or cystic fibrosis pathogenesis.
First, fragment libraries were constructed according to manufacturer’s instructions provided in the Ion Community for Ion TorrentTM sequencing using the panel and Ion AmpliSeqTM Library kit 2.0 (Life Technologies Inc., NY, USA).
Next, validation of enrichment and quantification of the resulting library were performed on the Agilent 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA, USA). Sample emulsion polymerase chain reaction, emulsion breaking, and enrichment were performed on Ion OneTouchTM (Life Technologies Inc.) using Ion OneTouchTM 200 Template Kit (Life Technologies Inc., NY, USA). Ion SphereTM Particles (ISPs) were recovered and template-positive ISPs were enriched using Dynabeads® MyOneTM Streptavidin C1 beads (Life Technologies Inc., NY, USA). ISP enrichment was confirmed using Qubit® 2.0 fluorometer (Life Technologies Inc., NY, USA), and the sample was prepared for sequencing using Ion PGMTM 200 Sequencing Kit protocol (Life Technologies Inc., NY, USA). The sample was then loaded on Ion 316TM chip and sequenced on Ion Torrent PGMTM (Life Technologies Inc., NY, USA). Softwares including Torrent SuiteTM 4.0 (Life Technologies INc., NY, USA), SeattleSeq Annotation 9.00 (Seattle, WA, USA) and Integrative Genomics Viewer 2.1 (IGV, Broad Institute of MIT and Harvard, Cambridge, MA, USA) were utilized to perform bioinformatics analysis, including optimized signal processing, base calling, sequence alignment, and variant analysis.
Finally, pathogenic variants were confirmed by Sanger sequencing using ABI 3730 DNA analyzer (Applied Biosystems, Foster City, CA, USA). Nomenclature for description of mutations was based on http://www.hgvs.org/mutnomen/, using NM_000138.4 as reference sequence.
Results
Mutants associated with CF
Eighteen CF mutants of 21 alleles were detected in 21 of bronchiectasis subjects. Of 100 healthy subjects, 9 CF mutants were detected in 11 cases (Table S4).
Acknowledgements
We thank the bronchiectasis patients who participated in our study and the staffs of the health check-up center for offering the blood specimens.
Funding: National Natural Science Foundation No. 81400010, Pearl River S&T Nova Program of Guangzhou No. 201710010097 and Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme 2017 (to Dr. Guan), National Natural Science Foundation No. 81500006 (to Dr. Gao), Changjiang Scholars and Innovative Research Team in University ITR0961, The National Key Technology R&D Program of the 12th National Five-year Development Plan 2012BAI05B01 and National Key Scientific & Technology Support Program: Collaborative innovation of Clinical Research for chronic obstructive pulmonary disease and lung cancer No. 2013BAI09B09 (to Profs. Zhong and Chen), and National Natural Science Foundation No. 81230015 and Beijing Science and Technology Plan No. Z151100003915078 (to Prof. Zhang).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: Ethics approval was obtained from Ethics Committee of The First Affiliated Hospital of Guangzhou Medical University. Written informed consent was obtained.
References
- Pasteur MC, Bilton D, Hill AT, et al. British Thoracic Society guidelines for non-CF bronchiectasis. Thorax 2010;65:i1-58. [Crossref] [PubMed]
- Pasteur MC, Helliwell SM, Houghton SJ, et al. An Investigation into Causative Factors in Patients with Bronchiectasis. Am J Respir Crit Care Med 2000;162:1277-84. [Crossref] [PubMed]
- Lonni S, Chalmers JD, Goeminne PC, et al. Etiology of Non-Cystic Fibrosis Bronchiectasis in Adults and Its Correlation to Disease Severity. Ann Am Thorac Soc 2015;12:1764-70. [Crossref] [PubMed]
- Guan WJ, Gao YH, Xu G, et al. Aetiology of bronchiectasis in Guangzhou, southern China. Respirology 2015;20:739-48. [Crossref] [PubMed]
- Davis SD, Ferkol TW, Rosenfeld M, et al. Clinical features of childhood primary ciliary dyskinesia by genotype and ultrastructural phenotype. Am J Respir Crit Care Med 2015;191:316-24. [Crossref] [PubMed]
- Knowles MR, Ostrowski LE, Leigh MW, et al. Mutations in RSPH1 cause primary ciliary dyskinesia with a unique clinical and ciliary phenotype. Am J Respir Crit Care Med 2014;189:707-17. [Crossref] [PubMed]
- Castleman VH, Romio L, Chodhari R, et al. Mutations in radial spoke head protein genes RSPH9 and RSPH4A cause primary ciliary dyskinesia with central-microtubular-pair abnormalities. Am J Hum Genet 2009;84:197-209. [Crossref] [PubMed]
- Panizzi JR, Becker-Heck A, Castleman VH, et al. CCDC103 mutations cause primary ciliary dyskinesia by disrupting assembly of ciliary dynein arms. Nat Genet 2012;44:714-9. [Crossref] [PubMed]
- Ardeniz Ö, Unger S, Onay H, et al. β2-Microglobulin deficiency causes a complex immunodeficiency of the innate and adaptive immune system. J Allergy Clin Immunol 2015;136:392-401. [Crossref] [PubMed]
- Liu Y, Wang L, Tian X, et al. Characterization of gene mutations and phenotypes of cystic fibrosis in Chinese patients. Respirology 2015;20:312-8. [Crossref] [PubMed]
- Lukowski SW, Bombieri C, Trezise AE. Disrupted post-transcriptional regulation of the cystic fibrosis transmembrane conductance regulator (CFTR) by a 5'UTR mutation is associated with a CFTR-related disease. Hum Mutat 2011;32:E2266-82. [Crossref] [PubMed]
- Aliberti S, Masefield S, Polverino E, et al. Research priorities in bronchiectasis: a consensus statement from the EMBARC Clinical Research Collaboration. Eur Respir J 2016;48:632-47. [Crossref] [PubMed]
- Schobel SA, Stucker KM, Moore ML, et al. Respiratory syncytial virus whole-genome sequencing identifies convergent evolution of sequence duplication in the C-terminus of the G gene. Sci Rep 2016;6:26311. [Crossref] [PubMed]
- Gao L, Bin L, Rafaels NM, et al. Targeted deep sequencing identifies rare loss-of-function variants in IFNGR1 for risk of atopic dermatitis complicated by eczema herpeticum. J Allergy Clin Immunol 2015;136:1591-600. [Crossref] [PubMed]
- Raje N, Soden S, Swanson D, et al. Utility of next generation sequencing in clinical primary immunodeficiencies. Curr Allergy Asthma Rep 2014;14:468. [Crossref] [PubMed]
- Kaderbhai CG, Boidot R, Beltjens F, et al. Use of dedicated gene panel sequencing using next generation sequencing to improve the personalized care of lung cancer. Oncotarget 2016;7:24860-70. [Crossref] [PubMed]
- Frommer A, Hjeji R, Loges NT, et al. Immunofluorescence analysis and diagnosis of primary ciliary dyskinesia with radial spoke defects. Am J Respir Cell Mol Biol 2015;53:563-73. [Crossref] [PubMed]
- Dougherty GW, Loges NT, Klinkenbusch JA, et al. DNAH11 localization in the proximal region of respiratory cilia defines distinct outer dynein arm complexes. Am J Respir Cell Mol Biol 2016;55:213-24. [Crossref] [PubMed]
- Guan WJ, Gao YH, Xu G, et al. Characterization of lung function impairment in adults with bronchiectasis. PLoS One 2014;9:e113373. [Crossref] [PubMed]
- Guan WJ, Gao YH, Xu G, et al. Sputum bacteriology in steady-state bronchiectasis in Guangzhou, China. Int J Tuberc Lung Dis 2015;19:610-9. [Crossref] [PubMed]
- Reiff DB, Wells AU, Carr DH, et al. CT findings in bronchiectasis: limited value in distinguishing between idiopathic and specific types. AJR Am J Roentgenol 1995;165:261-7. [Crossref] [PubMed]
- Chalmers JD, Goeminne P, Aliberti S, et al. The bronchiectasis severity index: An international derivation and validation study. Am J Respir Crit Care Med 2014;189:576-85. [Crossref] [PubMed]
- Hefferon TW, Groman JD, Yurk CE, et al. A variable dinucleotide repeat in the CFTR gene contributes to phenotype diversity by forming RNA secondary structures that alter splicing. Proc Natl Acad Sci U S A 2004;101:3504-9. [Crossref] [PubMed]
- Zielenski J, Patrizio P, Corey M, et al. CFTR gene variant for patients with congenital absence of vas deferens. Am J Hum Genet 1995;57:958-60. [PubMed]
- Chu CS, Trapnell BC, Curristin S, et al. Genetic basis of variable exon 9 skipping in cystic fibrosis transmembrane conductance regulator mRNA. Nature Genet 1993;3:151-6. [Crossref] [PubMed]
- Fajac I, Viel M, Sublemontier S, et al. Could a defective epithelial sodium channel lead to bronchiectasis. Respir Res 2008;9:46. [Crossref] [PubMed]
- Rademacher J, Schultz A, Hedtfeld S, et al. Nasal potential difference of carriers of the W304R ENaC variant with non-cystic fibrosis bronchiectasis. Eur Respir J 2016;47:322-4. [Crossref] [PubMed]
- Fajac I, Viel M, Gaitch N, et al. Combination of ENaC and CFTR mutations may predispose to cystic fibrosis-like disease. Eur Respir J 2009;34:772-3. [Crossref] [PubMed]
- Hornef N, Olbrich H, Horvath J, et al. DNAH5 mutations are a common cause of primary ciliary dyskinesia with outer dynein arm defects. Am J Respir Crit Care Med 2006;174:120-6. [Crossref] [PubMed]
- Cuppens H, Lin W, Jaspers M, et al. Polyvariant mutant cystic fibrosis transmembrane conductance regulator genes. The polymorphic (Tg)m locus explains the partial penetrance of the T5 polymorphism as a disease mutation. J Clin Invest 1998;101:487-96. [Crossref] [PubMed]
- Casals T, De-Gracia J, Gallego M, et al. Bronchiectasis in adult patients: an expression of heterozygosity for CFTR gene mutations? Clin Genet 2004;65:490. [Crossref] [PubMed]
- Hansson JH, Schild L, Lu Y, et al. A de novo missense mutation of the beta subunit of the epithelial sodium channel causes hypertension and Liddle syndrome, identifying a proline-rich segment critical for regulation of channel activity. Proc Natl Acad Sci U S A 1995;92:11495-9. [Crossref] [PubMed]
- Chang SS, Grunder S, Hanukoglu A, et al. Mutations in subunits of the epithelial sodium channel cause salt wasting with hyperkaliemic conditions acidosis, pseudohypoaldosteronism type 1. Nat Genet 1996;12:248-53. [Crossref] [PubMed]
- McShane PJ, Naureckas ET, Tino G, Strek ME. Non-cystic fibrosis bronchiectasis. Am J Respir Crit Care Med 2013;188:647-56. [Crossref] [PubMed]
- Ni Y, Shi G, Yu Y, et al. Clinical characteristics of patients with chronic obstructive pulmonary disease with comorbid bronchiectasis: a systemic review and meta-analysis. Int J Chron Obstruct Pulmon Dis 2015;10:1465-75. [Crossref] [PubMed]
- Lonni S, Chalmers JD, Goeminne PC, et al. Etiology of Non-Cystic Fibrosis Bronchiectasis in Adults and Its Correlation to Disease Severity. Ann Am Thorac Soc 2015;12:1764-70. [Crossref] [PubMed]
- Guan WJ, Yuan JJ, Gao YH, et al. Maximal mid-expiratory flow is a surrogate marker of lung clearance index for assessment of adults with bronchiectasis. Sci Rep 2016;6:28467. [Crossref] [PubMed]


