Neuregulin1 acts as a suppressor in human lung adenocarcinoma via AKT and ERK1/2 pathway
Introduction
Lung cancer originates from bronchial epithelial cells and is the leading cause of cancer-related deaths worldwide (1,2). Approximately, 1.83 million new cases of lung cancer were diagnosed in 2012 (http://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/lung-cancer), and about 733,000 newly diagnosed cases and 610,000 deaths were reported in 2015 in China (3). Presently, the treatment of lung cancer has a great limitation (4,5). However, recent studies have shown that phosphatidylinositol 3-kinase (PI3K) pathway and mitogen-activated protein kinase (MAPK) pathway play critical roles in the development of malignant tumors, including lung cancer (6-9). Thus, novel and effective treatment strategy is urgently requisite, the therapy targeted to the molecule related to the two pathways might be the potential approach.
Neuregulin1 (NRG1) is localized on the chromosome arm 8p, which is one of the most frequently altered regions in a large proportion of human cancers, including lung carcinoma (10-12). NRG1 gene encodes the NRG1 protein that belongs to the epidermal growth factor (EGF) family. It is expressed in various tissues and participates in their development and maturation (12). In addition, when coupled with the receptor tyrosine kinase (ERBBs) family, and activates the members it can serve as a signaling protein that is involved in several cell-cell signal transduction pathways including PI3K and MAPK pathways (13-15).
A number of studies have confirmed that NRG1 is abnormally expressed in a variety of tumors and associated with several aspects of tumor progression such as cell proliferation, differentiation, invasion, and metastasis (16-20). Also, it mediates the angiogenesis and alters the tumor cell morphology and tumor microenvironment (21-23).
Another study showed that NRG1 was overexpressed in lung cancer (16). Liu and Kern (18) confirmed that NRG1 promoted the proliferation of human lung adenocarcinoma (LUAD) cell line (NCI-H441) and human lung squamous cell carcinoma (LUSC) cell line (NCI-H520). Furthermore, blocking the signaling associated with NRG1 restrained the growth of primary non-small cell lung carcinomas (NSCLC) and enhanced the response to chemotherapy (24). Several investigators detected many kinds of gene fusions related to NRG1 including CD47-NRG1, SDC4-NRG1, RBPMS-NRG1, SLC3A2-NRG1, and WRN-NRG1 in lung cancer, which might be attributed to chromosomal rearrangements, interchromosomal translocations, or paracentric inversion in the arm of the corresponding region in the tumor cells (25-27). After the occurrence of gene fusion, the integrity of EGF structure in NRG1 was still retained that continued to persevere the biological effect (25,27).
However, only a few studies have addressed the clear relationship between lung cancer and NRG1, and only the present study has depicted the link between the two. Therefore, we hypothesized that NRG1 is expressed abnormally in LUAD tissues, thereby, affecting the biological behavior of the cell lines. The current research investigated the expression of NRG1 in LUAD tissues and analyzed the relationship between NRG1 expression and the clinical characteristics. Consecutively, the effects of NRG1 on the biological behaviors of human LUAD cell lines (A549 and H1975) and the potential mechanism of the functions were detected via systematic analysis on the role of NRG1 in human LUAD.
Methods
Patients and samples
The tissues of LUAD cancer and paracancer tissues were obtained from clinical specimens resected in the institution (Department of Cardiothoracic Surgery, Zhongnan Hospital) between September 2016 and April 2017 and confirmed by two independent pathologists. The patients, who provided specimens, neither presented any cardiovascular and nervous system diseases nor any primary tumor of other systems other than lung. Patients receiving neoadjuvant radiotherapy or chemotherapy preoperatively were excluded. The information on the clinical characteristics including age, gender, smoking, drinking history, tumor size, tumor differentiation, lymph node metastasis, and distant metastasis of relevant patients was collected. Specimens and clinical information were obtained with patients’ consent at the time of notification of surgical risks before operation. The study was approved by the Ethics Committee at Zhongnan Hospital of Wuhan University (No. 2015009), the samples collection, as well as treatment, was carried out in accordance with the Helsinki declaration guidelines.
Protein analyses
Isolation of total protein from LUAD tissues and LUAD cells
The fresh tissues were grounded with radio immunoprecipitation assay (RIPA) lysis buffer and protease inhibitors, or the LUAD cells (A549 and H1975) were sonicated and lysed in RIPA buffer containing protease inhibitor and phosphatase inhibitor. The homogeneous mixture was centrifuged at 12,000 ×g for 15 min to clarify the cell debris and nuclei. The total protein was collected from the supernatant and concentration estimated, followed by denaturation for 10 min in Laemmli buffer.
Western blot (WB) analysis
The respective total protein sample was resolved on 10% dodecyl sulfate, sodium salt (SDS)-polyacrylamide gel electrophoresis (PAGE) and transferred to a polyvinylidene fluoride (PVDF) membrane. After blocking in 5% nonfat dry milk in Tris buffered saline Tween (TBST) for 2 h, the membranes were probed appropriate primary antibodies at 4 °C overnight, followed by incubation with the corresponding secondary antibodies for 2 h at room temperature. Subsequently, the blots were developed by detection reagents, and immunoreactive bands quantified using Quantity One software. The primary antibodies included rabbit anti-human NRG1 polyclonal antibody (Abnove, Taiwan, Cat. #PAB4805, 1:1,000), rabbit anti-human AKT monoclonal antibody (Cell Signaling Technology, USA, Cat. #2938, 1:1,000), rabbit anti-human p-AKT polyclonal antibody (Abcam, USA, Cat. #ab23509, 1:1,000), rabbit anti-human ERK1/2 monoclonal antibody (CST, USA, Cat. #4695, 1:1,000), rabbit anti-human p-ERK1/2 monoclonal antibody (CST, USA, Cat. #4370, 1:2,000), and mouse anti-human β-Actin monoclonal antibody (Boster Biological Technology, Wuhan, Cat. #BM0627, 1:200).
mRNA analyses
Isolation of total RNA from LUAD tissues and LUAD cells
Total of 50 µg fresh tissues were homogenized with TRIzol (1 mL), or the LUAD cells (A549 and H1975) were sonicated and lysed in TRIzol. Then the total RNA was extracted with chloroform, isopropanol, and precooled ethanol, and estimated on a NanoDrop ND-1000 UV-Vis spectrophotometer (Thermo Scientific, USA).
Quantitative real-time polymerase chain reaction (qRT-PCR)
mRNA was reverse-transcribed to cDNA by the cDNA reverse transcriptase kit (Takara, Japan, Cat. #RR047A) according to the manufacturer’s instructions. The amplification of cDNA was carried out using the primers for β-Actin (forward: 5'-AGAGCTACGAGCTGCCTGAC-3', reverse: 5'-AGCACTGTGTTGGCGTACAG-3') and NRG1 (forward: 5'-AGTCCTTCGGTGTGAAACCAG-3', reverse: 5'-TGCGAAGTTCTGACTTCCCTG-3') on a Bio-Rad iCycler (USA, Cat. #CFX96). The reaction consisted of an activation step of 95 °C for 5 min, 40 cycles of 30 s at 95 °C, 30 s at 57 °C, and 45 s at 72 °C for denaturation, annealing, and extension, respectively, followed by final extension at 72 °C for 10 min. Each sample was amplified in triplicate, and the average Ct value of interest and internal reference gene for each sample was obtained for further analysis.
Immunohistochemistry (IHC)
The LUAD cancer tissue specimens were embedded in paraffin and sliced into thin sections (5 mm) after fixing in 4% formaldehyde for 24–36 h. Xylene, alcohol gradient, and distilled water were used deparaffinization of the sections, followed by the treatment with 3% H2O2 to block the endogenous peroxidase activity. Antigen retrieval was carried out by immersing the slides in sodium citrate. Non-specific Ig binding was blocked using 10% goat serum in phosphate-buffered saline (PBS) at a pH 7.4. The sections were incubated independently in rabbit anti-human NRG1 polyclonal antibody (Abnove, Cat. #PAB4805) at 4 °C for 12 h (1:50), followed by secondary antibody in the thermostat for 0.5 h. Subsequently, the sections were incubated with SABC (1:100) and DAB for color development, and the protein was counterstained with hematoxylin for 30 s, dehydrated with distilled water, and anhydrous alcohol. The slides were mounted using neutral gum. The protein was observed under a microscope, and the staining scored for expression intensity by two investigators. The final score equaled to the product of the score of the positive stain cell percentage and that of the intensity. The percentage of positive cell scored as follows: <1% scored 0 points, 1–25% scored 1 point, 26–50% scored 2 points, 51–75% scored 3 points, and >75% scored 4 points. The staining intensity (most cells showed prevalent positive staining) was scored as follows: no coloring scored 0, light tan scored 1 point, brown scored 2 points, dark brown scored 3 points. The total points 0–2 was considered to be negative (−), 3–4 was evaluated as weak positive (+), 5–7 was divided into moderate positive (++) and the scores ≥8 was strongly positive (+++).
Cell culture experiments on cell biological behavior
Cell culture and subculture
The human LUAD cell lines, A549 and H1975, were purchased from American Type Culture Collection (ATCC), and cultured in DMEM medium (Gibco, China) supplemented with 10% fetal bovine serum (FBS) (Evergreen, China), 1% penicillin-streptomycin sulfate in a humidified atmosphere consisting of 95% air and 5% CO2 at 37 °C. Subsequently, we sub-cultured when the cell density almost reached to 90%. The aseptic processing was essential during the whole cell experiments.
Knockdown of NRG1 in the LUAD cells
NRG1-target-specific siRNA (siRNA-NRG1) was synthesized by GenePharma (Suzhou, China). The sense sequence of siRNA-NRG1 was forward: 5'-GGUCUGAACGAAACAAUAUTT-3', reverse: 5'-AUAUUGUUUCGUUCAGACCTT-3'. According to the manufacturer’s protocol, siRNA-NRG1 and siRNA-control was transfected into LUAD cells (A549 and H1975), respectively, using lipofectamine 2000 (Invitrogen, USA). After transfection for 48 h, qRT-PCR, gel electrophoresis, and immunofluorescence were used to evaluate the alterations of NRG1 at the transcriptional level.
Concentration and duration of the activity of recombinant NRG1 (Re-NRG1)
The A549 cells, in logarithmic growth phase, were seeded in 96-well plants at a density of 2.5×103/well and cultured overnight in the presence of various, concentrations of Re-NRG1 (Abcam, USA, Cat. #Ab50227) at 0, 100, 200, 300, 400, 600, 800, and 1,200 ng/mL, respectively in five wells. Then, the plates were cultured for 12, 24, 48, and 72 h, respectively, followed by methylthiazolyldiphenyl-tetrazolium bromide (MTT) assay employed for the analysis of cell proliferation.
MTT assay on cell proliferation
After the cells were treated appropriately in 96-well plates, 20 µL MTT solution was added per well and incubated for 4 h for color development. Subsequently, the reaction was quenched with 100 µL dimethyl sulphoxide (DMSO), the liquid was removed after 15 min, and the absorbance was measured at OD568 using a microplate reader.
Flow cytometry assay on cell cycle and apoptosis
The flow cytometry experiments assessed the cell cycle and apoptosis of A549 and H1975. The cells, in the logarithmic growth phase, were seeded in 6-well plates at a density of 5×104/well. After appropriate treatments, the cells were harvested by trypsin without ethylene diamine tetraacetic acid (EDTA) for subsequent experiments of cell cycle and apoptosis. The cell cycle assay was performed using propidium iodide (PI) staining. Briefly, the trypsinized cells were collected and centrifuged at 1,200 ×g for 5 min and fixed with 70% ethanol at 4 °C for 4 h. Then the samples were incubated with RNase A (50 µg/mL DNase free) for 10 min at 37 °C, followed by 400 µL PI (50 µg/mL) at 4 °C for 15 min in the dark. Finally, the cell cycle test was assessed by flow cytometry. Cell apoptosis was assessed using the Annexin-V-FITC Apoptosis Detection Kit (Nanjing KeyGen Biotech, Inc, Cat. #KGA108) according to manufacturer’s instructions. Briefly, the digested cells were collected, centrifuged at 1,200 ×g for 5 min, and re-suspended in 500 µL PBS, 5 µL AnnexinV-FITC and PI were added into suspension, mixed well, and reacted for 10 min in the dark. Finally, the apoptosis was detected by flow cytometry. Normal cells showed FITC Annexin V (−) with PI (−) staining, early apoptotic was FITC Annexin V (+) with PI (−) staining, and late apoptotic was FITC Annexin V (+) with PI (+) or already dead cells stained with FITC Annexin V (−) and PI (+).
Transwell small chamber assay
The Transwell small chamber assay evaluated the cell invasion and migration of A549 and H1975 cells. The cell suspension containing 105 cells in the logarithmic growth phase was centrifuged at 1,500 ×g for 5 min. After mixing the cells with serum-free media 200 µL was added to the each Matrigel-containing 8 µm Transwell chamber. Then, the Transwell chambers were placed in 24-well plates with 10% complete medium. After respective treatments, the upper chambers were washed with PBS, the cells and Matrigel wiped off from the bottom chamber, and stained with crystal violet for 10 min. Subsequently, the images of side of the chamber without cultured cells were captured and the number of cells calculated. The cell migration assay was performed similarly, albeit without Matrigel.
Statistical analysis
The data were expressed as mean ± standard deviation (SD) from three independent experiments. Statistical analysis was performed using SPSS for Windows (version 17.0). The paired samples t-test and Fisher’s exact chi-square test evaluated the significance of statistical differences in the data between cell and tissue experiments. The significance of statistical differences of the data from cell experiments was evaluated by one-way analysis of variance (ANOVA) and Pearson’s chi-square test. A value of P<0.05 was considered statistically significant.
Results
NRG1 expression was downregulated in the cancer tissues of LUAD as compared to the paracancerous tissues
Forty-four pairs of LUAD cancer tissues and paracancerous tissues were used for the detection of NRG1 protein expression by WB. As shown in Figure 1A,B, NRG1 was expressed in both LUAD cancer and paired paracancerous tissues. Furthermore, compared to the expression in the two groups, the NRG1 expression in cancer tissues was lower than that in the paracancerous cancer tissues (P=0.039). Twenty-five pairs of LUAD cancer and paracancerous tissues were assessed for the mRNA expression of NRG1. The results (Figure 1C) demonstrated that both the LUAD cancer and paracancerous tissues expressed the mRNA of NRG1. Subsequently, 2−ΔCt was utilized for statistical analysis of the data, which showed that the LUAD cancer tissues expressed lower mRNA than the paracancerous tissues (P=0.024).

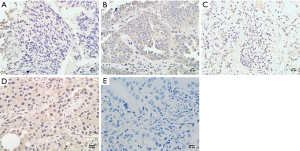
Expression of NRG1 in LUAD cancer tissues was related to clinical characteristics
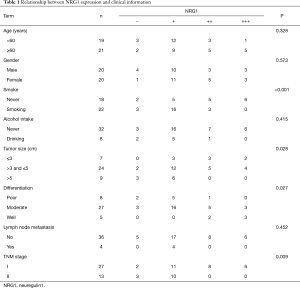
IHC was performed on tissue sections in 40 cases of LUAD tissues. The clinical data including the age, gender, history of smoking and alcohol intake, tumor size, differentiation degree, lymph node metastasis status, and distant metastasis were collected. The differential of protein expression was demonstrated by the different staining intensities and the different number of stained cells (Figure 2). The protein was primarily expressed in the nucleus; however, some cells also expressed it on the cell membranes and cytoplasm. The scoring of the above rating criteria and statistical analysis were illustrated in Table 1. Consequently, NRG1 expression was not associated with the patients’ age (P=0.328), gender (P=0.573), history of alcohol intake (P=0.415), and lymph node metastasis (P=0.452); however, it was related to the history of smoking (P<0.001), tumor size (P=0.028), degree of differentiation (P=0.027), and TNM stage (P=0.009).
Full table
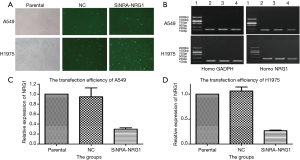
Transfection efficiencies of siRNA
After the cells were transfected and cultured for 48 h, followed by detection with immunofluorescence (Figure 3A). Total RNA was extracted from these cells to detect the efficiency of transfusion by gel electrophoresis and qRT-PCR (Figure 3B,C,D). The transfection efficiency was found to be 70.3% and 73.7% in A549 and H1975 cells, respectively.
Concentration and time for exogenous NRG1 activity.
The effects of seven different Re-NRG1 concentrations on the proliferation of A549 cell line at four time points were assessed by MTT assay (Figure 4A). The different doses of Re-NRG1 were not found to significantly affect the cell proliferation after it acted for 12, 24, and 48 h, respectively. However, after 72 h, the drug exerted an inhibitory effect on cell proliferation at the same dose. Also, the concentration at 300 ng/mL affected the cell proliferation maximally (P=0.001), and the maximum inhibitory rate was up to 26.9%. Beyond 300 ng/mL, the effect weakened gradually with increasing dosage. Furthermore, no difference was noted when the concentration reached 1,200 ng/mL (P=0.358). Therefore, in the subsequent experiments, the concentration for the activity of exogenous Re-NRG1 was set at 0, 300, and 1,200 ng/mL for 72 h.
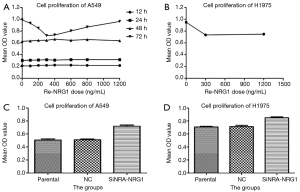
NRG1 inhibited the proliferation of LUAD cells
The activity of exogenous Re-NRG1 on H1975 cells at the concentration of 0, 300, 1,200 ng/mL for 72 h was detected based on cell proliferation using MTT assay (Figure 4B). The result showed that Re-NRG1 exerted made an inhibitory effect on cell proliferation (P<0.001), but not in a concentration-dependent manner (P=0.331).
After cells were transfected and cultured for 48 h, MTT assay was used for the detection of cell proliferation (Figure 4C,D). The results showed a facilitation effect on cell proliferation when NRG1 gene was knocked down to suppress the expression (P<0.001 in A549 and H1975 cells, respectively).
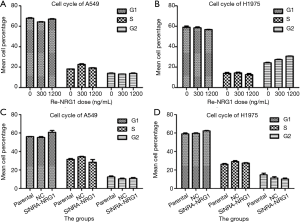
NRG1 did not affect cell cycle distribution
The effects of different doses of exogenous Re-NRG1 on the cell cycle distribution of A549 and H1975 cells were displayed in the Figure 5A,B. After cells were transfected and cultured for 48 h, the results of cell cycle distribution were illustrated in Figure 5C,D, which showed that NRG1 did not exhibit any difference on the cell cycle distribution under experimental conditions (P=0.753, 0.599, 0.532, 0.500).
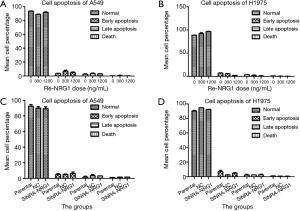
NRG1 did not effectuate a difference on the apoptosis status of LUAD cells
After up-regulation of NRG1 by addition of different doses into the LUAD cells (A549 and H1975), we adopted flow cytometry assay to detect the apoptosis status of both cells (Figure 6A,B); the results did not reveal any NRG1-mediated difference on the apoptosis status (P=0.753, 0.858). When the expression of NRG1 gene was downregulated by knockdown in the cells (Figure 6C,D), the apoptosis status was not found to be inhibited or promoted in A549 (P=0.831) and H1975 (P=0.332) cells.
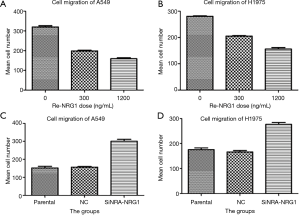
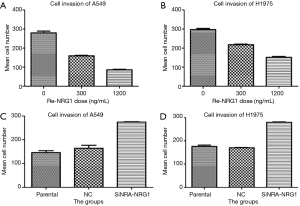
NRG1 inhibited cell invasion and migration ability
The role of different doses of exogenous Re-NRG1 on the migration and invasion of A549 and H1975 cells were shown in Figures 7,8. These displayed an inhibitory effect of NRG1 on the cell migration (P<0.001) and cell invasion (P<0.001). In addition, a strong inhibition occurred at high concentration. Moreover, when the expression of NRG1 was suppressed, the ability of cell invasion (P<0.001) and migration (P<0.001) was facilitated (Figures 7,8).
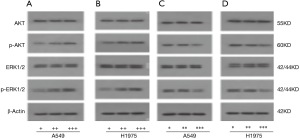
NRG1 played a role through AKT and ERK signaling pathways
The protein expression of AKT signaling pathway (t-AKT and p-AKT) and ERK signaling pathway (t-ERK and p-ERK) was detected. The results in Figure 9A,B demonstrated that when the level of NRG1 was upregulated, the expression of p-AKT and p-ERK increased (P<0.001, respectively). On the contrary, when the expression of NRG1 was downregulated by gene knockdown, the expression of p-AKT (P<0.001) and p-ERK (P<0.001) was promoted (Figure 9C,D). Consecutively, these results showed that the expression of t-AKT and t-ERK was not affected in either of the two experimental conditions.
Discussion
Lung cancer is a malignant tumor originating from bronchial epithelial cells, and a major concern with respect to high morbidity and mortality worldwide (1,2), which necessitates intensive focus for improvement in humans. As shown in the tumor molecular genetics and molecular biology, the tumorigenesis is caused by the activated oncogene or tumor suppressor gene (TSG) inactivated, especially the inactivation of TSG (28,29), in order to improve the survival rates and living standard of the lung cancer patients, investigating the pathogenesis of the malignant disease and identifying a new closely related target molecule might be an effective approach. A majority of the molecules associated with malignancies effectuate by activating or suppressing the signaling pathways such as MAPK and PI3K (6-9). Thus, the molecules associated with these pathways, such as NRG1, might play a critical role in lung cancer.
The NRG1 gene consists of 1.4 Mbp and is localized to the chromosome arm 8p, which is one of the most frequently altered regions in a variety of human cancers (10-12). NRG1 encodes the NRG1 protein that belongs to the EGF family (12); the protein plays a major role in the human nervous system and the cardiovascular system (30,31). The protein structure contains a distinct region known as the EGF-like domain that is located in the membrane-proximal region of the extracellular area and is vital in activating the receptor ERBB members for the biological roles as a signaling protein involved in several cell-cell signaling transduction pathways (12,32). Various studies indicated that NRG1 overexpressed in several human tumor tissues and promoted the processing of tumor proliferation, differentiation, migration, angiogenesis, and the change in the microenvironment (16-23) by combining with the members of ERBB family to activate the multiple downstream signaling pathways such as MAPK and PI3K (14,15). These studies proved that NRG1 acts as a promotor oncogene in cancer development. Conversely, Chua et al. (33) observed that NRG1 expression was reduced in various breast cancer cell lines due to gene methylation. Additional studies confirmed the loss of NRG1 gene, which resulted from the chromosomal abnormality that was related to the occurrence of several tumors: breast cancer and colon cancer (10,11). Thus, the NRG1 is a candidate for both oncogene and suppressor gene (34,35) depending on the type of cancers, and hence, the role of NRG1 in malignant tumors is yet controversial.
Some studies found that NRG1 was overexpressed in lung cancer (16) and blocking the signalling associated with NRG1 could restrain the growth of primary NSCLC and enhanced the response to chemotherapy (24). Fernandez-Cuesta et al. (25,27) found CD74-NRG1 gene fusion in 22 invasive mucinous adenocarcinomas, while the SLC3A2-NRG1 gene fusion was found by Nakaoku et al. (26) in this tissue. Dhanasekaran et al. (4) observed four kinds of gene fusions related to NRG1 including CD74-NRG1, SDC4-NRG1, RBPMS-NRG1, and WRN-NRG1 in 733 lung cancer cases by RNA sequencing. Further studies have shown that the occurrence of these gene fusions may be due to the chromosomal rearrangements, chromosomal translocation, or chromosomal inversion in the arm of the corresponding region in the tumor cells (25,26). Subsequently, the ability to express the EGF structure was still retained completely (25,27), and hence, the main function of NRG1 might also lead to the inactivation of some tumor suppressor. In summary, NRG1 may play a critical role in lung cancer, and further investigation may provide a novel theoretical basis for lung cancer treatment.
In the current study, we found that NRG1 expressed in both the LUAD cancer and paracancerous tissues, and the expression was lower in cancer than the paracancerous tissues. Also, we concluded that the lower NRG1 expression in the cancerous tissues was negatively related to poor differentiation, small diameter, and early TNM stage, which differed from the study that pointed out that the expression of NRG1 in LUAD was only gender-related and independent of other clinical features (36). However, the study did not address the differential expression of NRG1 in cancer and paracancerous tissues, and herein, we did not detect the relationship between NRG1 expression and the survival time. Therefore, NRG1 might be associated with LUAD. Furthermore, we also upregulated and downregulated the expression of NRG1, and proved that NRG1 inhibited the proliferation of A549 and H1975 simultaneously; however, the maximum inhibition rate was only 26.9% and 22.72%, respectively, which indicated that the Re-NRG1 inhibited only the growth and did not induce cell death. Liu and Kern (18) showed that NRG1 promoted the proliferation of LUAD cell line, NCI-H441, while the current result was contradictory. Nevertheless, the two studies used different cell lines that might ascribe to different conclusions. In our current studies, the invasion and migration ability were also reduced under the influence of upregulated NRG1 and stimulated when NRG1 was downregulated. Thus, NRG1 may exert a protective role in the metastasis of lung cancer. Further analysis of the experimental results regarding the change in the biological behaviour under the influence of NRG1 can be identical with the studies about gene expression in the tissues. We did not observe any effect on the cell cycle and apoptosis, which might be attributed to insufficient dose or short duration of the reaction.
AKT is a serine/threonine protein kinase in the PI3K signal transduction pathway, and the abnormal activation occurs in a variety of malignant tumors (6). The ERK1/2 pathway plays a critical role in cell proliferation, differentiation, and survival and is one of the major pathways in the MAPK pathway (37). A number of studies have proved that PI3K and MAPK pathways exert vital roles in the development of malignant tumors, including lung cancer (6-9). We detected the expression of AKT and EKT1/2 pathway proteins after upregulation of NRG1 post-exposure of the cells to exogenous Re-NRG1, as well as after downregulated of NRG1 by gene knockdown. Consequently, the expression of t-AKT and t-ERK1/2 was unaltered in these experimental conditions. However, the expression of p-AKT and p-ERK1/2 was reduced when NRG1 was downregulated, while it was increased after NRG1 upregulation. Based on a comprehensive analysis, we can speculate that the activation of AKT and ERK1/2 pathway is closely related to the role of NRG1 in LUAD.
Conclusions
In conclusion, NRG1 decreased the expression in LUAD tissues and inhibited some biological behaviors of A549 and H1975 cell lines. Thus, NRG1 is speculated to plays a role in the development of LUAD as a TSG via AKT and ERK1/2 pathways. However, since only a small number of tissue samples and limited cell lines were included in the study, further research is essential.
Acknowledgements
The excellent technical assistance of Yuming Cao, Hong Cheng, Yuquan Bai is gratefully acknowledge. We would like to thank Department of Cardiothoracic Surgery, Zhongnan Hospital for equipment support.
Funding: This work was supported by the grant from The Key Project of Hubei Natural Science Foundation of China (No. 2011CDA064) and Hubei Province Natural Science Fund Project to Yuting Bai. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by the Ethics Committee at Zhongnan Hospital of Wuhan University (No. 2015009) and written informed consent was obtained from all patients.
References
- Siegel R, Naishadham D, Jemal A. Cancer Statistics 2013. CA Cancer J Clin 2013;63:11-30. [Crossref] [PubMed]
- Jemal A, Bray F, Center MM, et al. Global Cancer statistics CA Cancer J Clin 2011;61:69-90. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Dhanasekaran SM, Balbin OA, Chen G, et al. Transcriptome meta-analysis of lung cancer reveals recurrent aberrations in NRG1 and Hippo pathway genes. Nat Commun 2014;5:5893. [Crossref] [PubMed]
- Lemjabbar-Alaoui H, Hassan OU, Yang YW, et al. Lung cancer: Biology and treatment options. Biochim Biophys Acta 2015;1856:189-210. [PubMed]
- Martelli AM, Tabellini G, Bressanin D, et al. The emerging multiple roles of nuclear Akt. Biochim Biophys Acta 2012;1823:2168-78. [Crossref] [PubMed]
- Fei HR, Chen G, Wang JM, et al. Perifosine induces cell cycle arrest and apoptosis in human hepatocellular carcinoma cell lines by blockade of Akt phosphorylation. Cytotechnology 2010;62:449-60. [Crossref] [PubMed]
- Zhang W, Liu HT. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res 2002;12:9-18. [Crossref] [PubMed]
- Kitano H, Chung JY, Ylaya K, et al. Profiling of phospho-AKT, phospho-mTOR, phospho-MAPK and EGFR in non-small cell lung cancer. J Histochem Cytochem 2014;62:335-46. [Crossref] [PubMed]
- Birnbaum D, Adelaide J, Popovici C, et al. Chromosome arm 8p and cancer: a fragile hypothesis. Lancet Oncol 2003;4:639-42. [Crossref] [PubMed]
- Pole JC, Courtay-Cahen C, Garcia MJ, et al. High-resolution analysis of chromosome rearrangements on 8p in breast, colon and pancreatic cancer reveals a complex pattern of loss, gain and translocation. Oncogene 2006;25:5693-706. [Crossref] [PubMed]
- Falls DL. Neuregulins: functions, forms, and signaling strategies. Exp Cell Res 2003;284:14-30. [Crossref] [PubMed]
- Breuleux M, Schoumacher F, Rehn D, et al. Heregulins Implicated in Cellular Functions Other Than Receptor Activation. Mol Cancer Res 2006;4:27-37. [Crossref] [PubMed]
- Amin DN, Tuck D, Stern DF. Neuregulin-regulated gene expression in mammary carcinoma cells. Exp Cell Res 2005;309:12-23. [Crossref] [PubMed]
- Tsai MS, Shamon-Taylor LA, Mehmi I, et al. Blockage of heregulin expression inhibits tumorigenicity and metastasis of breast cancer. Oncogene 2003;22:761-8. [Crossref] [PubMed]
- Meetze K, Vincent S, Tyler S, et al. Neuregulin 1 Expression Is a Predictive Biomarker for Response to AV-203, an ERBB3 Inhibitory Antibody, in Human Tumor Models. Clin Cancer Res 2015;21:1106-14. [Crossref] [PubMed]
- Montero JC, Rodriguez-Barrueco R, Ocana A, et al. Neuregulins and Cancer. Clin Cancer Res 2008;14:3237-41. [Crossref] [PubMed]
- Liu J, Kern JA. Neuregulin-1 Activates the JAK-STAT Pathway and Regulates Lung Epithelial Cell Proliferation. Am J Respir Cell Mol Biol 2002;27:306-13. [Crossref] [PubMed]
- Chen Q, Zhang NL, Gray RS, et al. A temporal requirement for Hippo signaling in mammary gland differentiation, growth, and tumorigenesis. Genes Dev 2014;28:432-7. [Crossref] [PubMed]
- Haskins JW, Nguyen DX, Stern DF. Neuregulin 1-activated ERBB4 interacts with YAP to induce Hippo pathway target genes and promote cell migration. Sci Signal 2014;7:ra116. [Crossref] [PubMed]
- Momeny M, Saunus JM, Marturana F, et al. Heregulin-HER3-HER2 signaling promotes matrix metalloproteinase-dependent blood-brain-barrier transendothelial migration of human breast cancer cell lines. Oncotarget 2015;6:3932-46. [Crossref] [PubMed]
- Asrani K, Keri RA, Galisteo R, et al. The HER2- and Heregulin 1 (HRG)-Inducible TNFR Superfamily Member Fn14 Promotes HRG-Driven Breast Cancer Cell Migration, Invasion, and MMP9 Expression. Mol Cancer Res 2013;11:393-404. [Crossref] [PubMed]
- Yen L, You XL, Al Moustafa AE, et al. Heregulin selectively upregulates vascular endothelial growth factor secretion in cancer cells and stimulates angiogenesis. Oncogene 2000;19:3460-9. [Crossref] [PubMed]
- Hegde GV, de la Cruz CC, Chiu C, et al. Bloking NRG1 and other ligand-mediated Her4 signaling enhances the magnitude and duration of the chemotherapeutic response of non-small cell lung cancer. Sci Transl Med 2013;5. [Crossref] [PubMed]
- Fernandez-Cuesta L, Plenker D, Osada H, et al. CD74-NRG1 Fusions in Lung Adenocarcinoma. Cancer Discov 2014;4:415-22. [Crossref] [PubMed]
- Nakaoku T, Tsuta K, Ichikawa H, et al. Druggable Oncogene Fusions in Invasive Mucinous Lung Adenocarcinoma. Clin Cancer Res 2014;20:3087-93. [Crossref] [PubMed]
- Fernandez-Cuesta L, Thomas RK. Molecular Pathways: Targeting NRG1 Fusions in Lung Cancer. Clin Cancer Res 2015;21:1989-94. [Crossref] [PubMed]
- Sanchez-Tapia C, Wan FY. Fastest time to cancer by loss of tumor suppressor genes. Bull Math Biol 2014;76:2737-84. [Crossref] [PubMed]
- Morris LG, Chan TA. Therapeutic targeting of tumor suppressor genes. Cancer 2015;121:1357-68. [Crossref] [PubMed]
- Esper RM, Pankonin MS, Loeb JA. Neuregulins: versatile growth and differentiation factors in nervous system development and human disease. Brain Res Rev 2006;51:161-75. [Crossref] [PubMed]
- Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol 2001;2:127-37. [Crossref] [PubMed]
- Mei L, Xiong WC. Neuregulin 1 in neural development, synaptic plasticity and schizophrenia. Nat Rev Neurosci 2008;9:437-52. [Crossref] [PubMed]
- Chua YL, Ito Y, Pole JC, et al. The NRG1 gene is frequently silenced by methylation in breast cancers and is a strong candidate for the 8p tumour suppressor gene. Oncogene 2009;28:4041-52. [Crossref] [PubMed]
- Korkaya H, Paulson A, Iovino F, et al. HER2 regulates the mammary stem/progenitor cell population driving tumorigenesis and invasion. Oncogene 2008;27:6120-30. [Crossref] [PubMed]
- Weinstein EJ, Grimm S, Leder P, et al. The oncogene heregulin induces apoptosis in breast epithelial cells and tumors. Oncogene 1998;17:2107-13. [Crossref] [PubMed]
- Pan B, Wang R, Huang Y, et al. HGF and NRG1 protein expression are not poor prognostic markers in surgically resected lung adenocarcinoma. Onco Targets Ther 2015;8:1185-91. [Crossref] [PubMed]
- Chye SM, Tiong YL, Yip WK, et al. Apoptosis induced by para-phenylenediamine involves formation of ROS and activation of p38 and JNK in chang liver cells. Environ Toxicol 2014;29:981-90. [Crossref] [PubMed]


