
Mutational profile of colorectal cancer lung metastases and paired primary tumors by targeted next generation sequencing: implications on clinical outcome after surgery
Introduction
Currently 1.4 million people are newly diagnosed with colorectal cancer (CRC) per year, making it one of the most common malignancies worldwide (1). By 2030, an increase of the incidence and mortality rate of 60% is expected (2). Disease spread to distant organs is considered the main reason for morbidity and mortality in patients with CRC (3). The lungs—owing its extensive microvascular network and favorable microenvironment—belong to the organs typically affected by metastatic dissemination. Possible treatment modalities for CRC pulmonary metastases (PM) comprise chemotherapy, radiation therapy and surgery or a combination thereof. Pulmonary metastasectomy can provide reasonable long-term survival, even in the setting of repeated metastasectomy (4-6). The selection criteria for pulmonary metastasectomy are currently based on technical considerations, i.e., complete resectability, and the functional status of the patient. Clinical prognostic variables reflecting an aggressive tumor biology such as multiple metastasis or a short disease-free interval (DFI) between the primary tumor and PM have been extensively described in the literature. However, these risk factors are hardly used in the clinical practice. Thus, there is a need for novel prognostic markers, enabling the surgeon to identify patients who will benefit from an invasive surgical procedure like pulmonary metastasectomy (7-9).
Despite recent advances in understanding the CRC tumor biology, it is not used as decision criteria when selecting treatment options for patients with PM (10). Since the English surgeon Stephen Paget [1855–1926] established the “seed and soil” hypothesis, a main goal of cancer research has been to decipher the clinical course of a malignant disease by analyzing the underlying tumor biology (11). Extensively studied genes in CRC include the KRAS and BRAF genes (12). However, given the great variety of genetic alterations in CRC, more elaborated and comprehensive analytical tools might allow further insights into the metastatic process of CRC and impact clinical decision-making.
Next-generation sequencing (NGS) has become an affordable and powerful tool to assess complete mutational profiles of cancer patients (13,14). In combination with large databases and advanced statistical methods, NGS could provide an individualized treatment to cancer patients.
In this study we aimed to describe the mutational profile of PM and corresponding primary CRC of patients treated in a single institution using targeted NGS (tNGS) approach. Moreover, advanced statistics were used to identify candidate genes, which impact the clinical outcome after pulmonary metastasectomy.
Methods
Study population
From April 2009 to June 2014, 57 patients with primary CRC received macroscopically and microscopically complete pulmonary metastasectomy at the Division of Thoracic Surgery, Medical University of Vienna. Clinical data based on a prospectively fed institutional database was retrospectively analyzed. Periodical computed tomography scans (3, 6, 9 and 12 months after surgery, thereafter biannually) were conducted during the follow-up after metastasectomy. Next generation sequencing of PM was successful in 47/57 (82.5%) patients. In addition, sequencing data was obtained from 24/31 (77.4%) corresponding primary tumors. The study was approved by the institutional review board (#1044/2012) and conducted according the Declaration of Helsinki.
tNGS
DNA was isolated from formalin-fixed paraffin-embedded (FFPE) tumor tissue samples using QiaAmp FFPE Tissue Kit (Qiagen) according to the manual provided by the manufacturer. DNA quantification was performed using Qubit® dsDNA HS Assay Kit (ThermoFisher Scientific). A qPCR-based Illumina quality control (QC) assay was used to confirm sufficient DNA quality for subsequent NGS.
DNA samples were subjected to NGS library preparation using TruSeq Amplicon Cancer Panel provided by Illumina (San Diego, CA, USA), which allows parallel investigation of 48 cancer-relevant genes. Pooled Libraries were sequenced with MiSeq Illumina platform using MiSeq v2 (300 cycles) sequencing chemistry. Primary data analysis was performed via Basespace Sequence Hub using TruSeq Amplicon pipeline (version 2.0.0.0. Alignment to hg19 reference genome using SAMtools Isis Smith-Waterman-Gotoh and base calling with somatic variant caller). For primary filtering and variant annotation of present dataset Illumina Variantstudio v2.2 software was used. Combined datasets were filtered manually according to predefined criteria. Regarding the effect of a certain variant, 3' and 5' prime UTR variants, intron variants, synonymous and non-coding exon variants were excluded, respectively.
Statistical analysis
The DFI was defined as the time between surgery for the primary tumor and pulmonary metastasectomy in months. Time to recurrence (TTR) represented the time between pulmonary metastasectomy and evidence of recurrence at any organ site. Overall survival (OS) was defined as the period of time between pulmonary metastasectomy and death of any cause. If patients had a history of pulmonary metastasectomy before the inclusion period, the previously resected metastases were assessed and outcome was calculated from the first pulmonary metastasectomy. Nominal variables were compared using Fisher’s exact test and chi-square test. Benjamini-Hochberg correction was performed if necessary. Survival curves were estimated using Kaplan-Meier plots and the difference of the groups were compared using the log-rank test. Statistics were performed using SPSS 23 (SPSS Inc., Chicago, USA) and GraphPad Prism 6 (GraphPad Software Inc., California, USA) software. All performed tests were two-sided. P values ≤0.05 were considered statistically significant.
Twenty-eight genes with more than six mutations from all 47 patients (ABL1, APC, ATM, CDH1, EGFR, ERBB4, FBXW7, FGFR2, FLT3, GNA11, GNAQ, HNF1A, HRAS, JAK3, KDR, KIT, KRAS, MET, NRAS, PDGFRA, PIK3CA, PTEN, PTPN11, RB1, RET, SMAD4, SMARCB1, and TP53) were used for regularized elastic-net Cox regression analyses (hdnom v4.8.9000, R-package) (15), together with a selection of clinicopathologic parameters, number of PM, age, sex, primary T-stage, primary N-stage, UICC-stage, previous liver metastases, DFI between primary tumor and lung metastasis and location of the primary tumor.
Outcomes were either censored OS or time to pulmonary recurrence (TTPR) data. Optimal lambda values were determined by internal N-fold cross-validation evaluating the penalized partial likelihood. Models are presented in nomograms. Performance of the final OS and TTPR adaptive elastic-net models were validated by bootstrap resampling, at every half year from the first (TTPR) or second (OS) year to the third (TTPR) or sixth (OS) year. Mean (solid line) and median (dashed line) area under the receiver operating characteristic (ROC) curve (AUC) values at each time point with the 25th to 75th percentile range (dark grey) and the total range (grey) are shown. Finally, the TTPR and the OS models were dichotomized into two equally large low- and high-risk groups and survival estimates shown in plots. As these are survival estimates from multiple Cox-regression models no censored patients are shown as in Kaplan-Meier estimates.
Results
Forty-seven patients with histologically verified PM from primary CRC were included in this study. Median age of the patients was 63 years (range, 44–83 years), 26 (55.3%) were male and 21 (44.7%) female. Twenty-eight (59.6%) of the primary tumors were located in the colon and the remaining 19 (40.4%) originated from the rectum. Detailed baseline characteristics of the study cohort are provided in Table 1.

Full table
Mutation frequency in PM and corresponding primary tumors
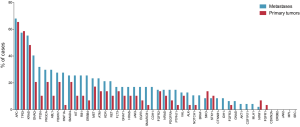
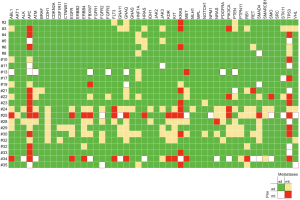
Mutations were most frequently found in APC, TP53 and KRAS with rates of 65.5% and 68.1% (APC), 58.6% and 57.4% (TP53), 48.3% and 55.3% (KRAS) in primary tumors and corresponding PM, respectively. CDH1, BRAF, CTNNB1, IDH1, GNAS, AKT1, CSFR11 and MLH1 mutations were exclusively found in PM, whereas FGFR1 was solely evident in primary tumors. In general, mutation frequencies were higher in PM compared to primary CRC. Mutation frequencies of all 48 assessed genes are depicted in Figure 1. When comparing the subgroup of metastases for which a matched primary was available, identical genotype was found in 83.5% (both non-mutated 78.2%; both mutated 5.3%, respectively). A new mutation in metastatic tissue compared to wildtype primary tumors were found in 12.8% of assessed genes. In 3.7%, mutations were found in the primary tumor, but could not be detected in the corresponding PM (Figure 2). The frequency of mutated genes was comparable between patients with and without chemotherapy before metastasectomy (Benjamini-Hochberg corrected Fisher’s exact P=n.s.).
Outcome analysis
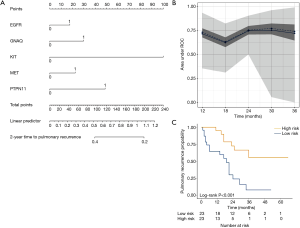
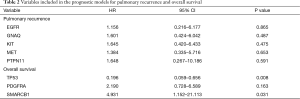
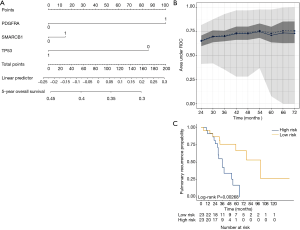
We aimed to assess the prognostic impact of the mutational profile on clinical outcome after surgery, defined by the time to pulmonary recurrence (TTPR) and OS. Median time of follow-up was 33 months. Adaptive elastic-net regularized Cox-regression models were built from mutation data and selected clinicopathologic data. The resulting multivariate model included the mutation status of EGFR, GNAQ, KIT, MET and PTPN11, all independently predicting TTPR. Noteworthy, mutations in KIT and PTPN11 had the strongest prognostic impact (Figure 3 and Table 2). Furthermore, a model comprising the mutational status of SMARCB1, PDGFRA and TP53 reflected the patients’ OS after pulmonary metastasectomy (Figure 4). PDGFRA mutations had the strongest negative impact on the OS in our model.
Full table
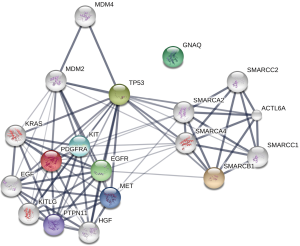
Pathway enrichment analysis of identified prognostic genes
Next, a pathway enrichment analysis using the STRING database was performed including the prognostic genes from the previous outcome analysis. EGFR, GNAQ, KIT, MET, PTPN11, PDGFRA, SMARCB1 and TP53 were included. This method facilitates the visualization of relevant signaling pathways. The STRING database highlights protein-protein interactions based on evidence from published literature (16). A significant interaction between the outcome relevant genes could be found (Figure 5; P=0.007). The two strongest affected pathways were the RAS signaling pathway (EGFR, KIT, MET and PTPN11) and the RAP1 signaling pathway (EGFR, GNAQ, KIT and MET). A detailed list of relevant pathways is provided in Table S1.

Full table
Discussion
This study aimed to comprehensively characterize the mutational profile of patients with metastatic CRC undergoing pulmonary metastasectomy. We were able to identify prognostic gene signatures and signaling pathways relevant in the metastatic process of CRC lung metastasis. In addition, we could define mutation signatures associated with impaired prognosis. To the best of our knowledge this is the first work applying a tNGS approach combined with advanced statistics in a considerable number of patients.
In patients with PM, resection of metastatic nodules is a widely applied treatment strategy. Despite the great number of patients receiving pulmonary metastasectomy, little is known about the tumor biology of these patients. With the emergence of targeted therapies, the identification of specific alterations at DNA, RNA and protein level has become of increasing importance. Moreover, the impact of these alterations on the management of patients receiving surgery is currently unknown. In CRC liver metastases, recent effort has been made to identify prognostic relevant mutations. The most prevalent mutated gene is KRAS (25–52%), which seems of prognostic relevance. However, the strongest negative impact on the prognosis is conferred by BRAF mutant tumors, which occur more rarely (0–9.1%) (17). So far, little is known about the genetic differences between CRC liver and lung metastases. However, Tie et al. found—based on a 19-gene array—significant differences between lung, brain and liver metastases. Thus, the underlying tumor biology and key mutations might differ between various sites of metastatic manifestation (18). Biomarkers reflecting the tumor biology might have an impact on the aggressiveness of surgical (re-)interventions by means of patient selection. Most thoracic surgeons are reluctant to offer pulmonary metastasectomy to patients with an aggressive tumor phenotype. In these cases, a removal of PM is frequently challenged by an early tumor recurrence, thus exposing patients mainly to the risks of a surgical procedure without a clear oncological benefit. On the other hand, pulmonary metastasectomy can be a curative treatment for patients with oligometastatic disease and a favorable tumor biology.
In addition, molecular profiling of metastatic tissue might impact the type of administered chemotherapy in the near future. A comprehensive analysis of the mutational profile specifically in patients with PM from CRC becomes increasingly important for surgical oncology. The prevalence of mutations in our cohort is comparable to previous reports in CRC, with APC, TP53 and KRAS being the most commonly mutated genes (19-21). Overall concordance of mutated genes between primary tumors and PM was 83.5%, which is similar to the concordance of 78% described by Vignot et al. in patients with CRC liver metastasis (22). Interestingly, we found 147 de novo mutations evident in lung metastases, which were not detected in the corresponding primary tumor. In contrast to this, 43 mutations were detected in the primary tumor but were not anymore evident in the matched PM. These findings have therapeutic and prognostic implications. First, targeted therapies, which have been proven beneficial in primary tumors need to be reevaluated in the metastatic setting. Additional diagnostic sampling of the distant metastases might help to find optimal treatment options. Second, our finding underlines clinical observations of usually aggressive distant spread, even in low grade primary tumors.
Furthermore, we could establish an internally cross-validated model of mutations, predicting TTPR and OS. Mutations in EGFR, GNAQ, KIT, MET and PTPN11 genes were associated with an early pulmonary recurrence after the first pulmonary metastasectomy. Interestingly, all five genes are linked to the RAS and RAP1 signaling pathways. Several groups have recently demonstrated the prognostic relevance of activating KRAS mutations in patients with CRC lung metastases (23-26). Herein, we could show that also other genetic alterations of this pathway are relevant in the process of metastasis in CRC. In addition to RAS, the RAP1 pathway was frequently affected in our study cohort. RAP1 plays a crucial role in cell-cell adhesion and cell-matrix interactions regulating adhesion molecules such as cadherins and integrins (27). It is not surprising, that altered genes incorporated in this pathway (EGFR, GNAQ, KIT, MET and PDGFRA) seem to be highly relevant in patients with lung metastases from CRC.
Three genes were identified to be of prognostic importance in the model for OS after pulmonary metastasectomy (PDGFRA, TP53 and SMARCB1) with wildtype TP53 being associated with an impaired prognosis. The prognostic impact of TP53 mutations are controversially discussed in the literature. Whereas in the majority of cancers, tumors harboring an altered TP53 have an aggressive behavior with early metastasis and poor OS, its role in CRC is unclear. Dallol et al. reported a worse prognosis of patients with KRAS mutated CRC when no mutation could be found in the TP53 gene (28). This unexpected observation was also made in the setting of CRC liver metastasectomy (29). Thus, TP53 independent tumor progression seems to be clinically more relevant than mutations in TP53.
PDGFRA, a receptor tyrosine kinase located on the cell surface, mediates several tumor-promoting effects after dimerization and activation by platelet-derived growth factor (PDGF)-isoforms. In CRC, PDGFRA overexpression is associated with advanced disease and lymph node involvement (30). Moreover, mutations identified by NGS have recently been described in therapy refractory CRC (31).
SMARCB1 is a tumor suppressor gene playing a role in chromatin remodeling. On protein level it is known to be relevant in the metastatic process of CRC and a loss of expression is associated with worse outcome (32,33). In contrast to our study, performing targeted NGS in 112 samples of primary CRC, Jauhri et al. detected a SMARCB1 mutation only in one patient (34). Kovaleva et al. described de novo SMARCB1 mutations in 2/14 (14%) examined CRC lung metastases (21). This frequency is comparable to our cohort, in which SMARCB1 mutations were detected in 8 of 47 samples.
There are several limitations to this study. First, due to its retrospective nature, only FFPE samples were available. Hence, the quality of the analyzed DNA might be reduced by fixation-induced artifacts when compared to fresh-frozen samples (35). Indeed, only 80.7% (71/88) of available samples had a sufficient quality to perform tNGS. However, usage of FFPE material reflects the situation in the clinical setting adequately, as only FFPE material will be available for most patients. Furthermore, patients analyzed in this study are a highly selected subgroup of patients with metastatic CRC, eligible for surgery. Thus, a selection bias is inevitably present. An extrapolation of our results to the total population of patients with metastatic CRC is therefore not possible. Moreover, patients did not receive a uniform chemotherapy before metastasectomy. Therefore, heterogeneity in the clonal selection of tumor cells by chemotherapeutic agents might have influenced the sampled tumors in our study. Precaution is warranted in the interpretation of the obtained data. Recommendations on patient selection or management cannot be derived from this study due to the limited number of patients. Confirmation in an external validation cohort will be required.
In summary, we herein describe a cross-validated prognostic model based on mutations identified by tNGS. These findings might have an impact on the selection and treatment of future patients with CRC lung metastasis. Further studies are warranted to confirm our results in a larger study cohort.
Acknowledgements
Funding: This study was supported by a research grant (#15880) provided by the Austrian Federal Bank (OeNB) and the Medical Scientific Fund of the Mayor of the City of Vienna (#15086).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by the institutional review board (#1044/2012) and conducted according the Declaration of Helsinki.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Arnold M, Sierra MS, Laversanne M, et al. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017;66:683-91. [Crossref] [PubMed]
- Mitry E, Guiu B, Cosconea S, et al. Epidemiology, management and prognosis of colorectal cancer with lung metastases: a 30-year population-based study. Gut 2010;59:1383-8. [Crossref] [PubMed]
- Kandioler D, Kromer E, Tuchler H, et al. Long-term results after repeated surgical removal of pulmonary metastases. Ann Thorac Surg 1998;65:909-12. [Crossref] [PubMed]
- Welter S, Jacobs J, Krbek T, et al. Long-term survival after repeated resection of pulmonary metastases from colorectal cancer. Ann Thorac Surg 2007;84:203-10. [Crossref] [PubMed]
- Dickinson KJ, Blackmon SH. Results of Pulmonary Resection: Colorectal Carcinoma. Thorac Surg Clin 2016;26:41-7. [Crossref] [PubMed]
- Lang C, Hrdliczka E, Schweiger T, et al. Impact of cyclooxygenase-2 and prostaglandin-E2 expression on clinical outcome after pulmonary metastasectomy. J Thorac Dis 2017;9:621-35. [Crossref] [PubMed]
- Hashimoto M, Tanaka F, Yoneda K, et al. The clinical value of circulating tumour cells (CTCs) in patients undergoing pulmonary metastasectomy for metastatic colorectal cancer. J Thorac Dis 2018;10:1569-77. [Crossref] [PubMed]
- Ampollini L, Gnetti L, Goldoni M, et al. Pulmonary metastasectomy for colorectal cancer: analysis of prognostic factors affecting survival. J Thorac Dis 2017;9:S1282-90. [Crossref] [PubMed]
- Schweiger T, Lang G, Klepetko W, et al. Prognostic factors in pulmonary metastasectomy: spotlight on molecular and radiological markers. Eur J Cardiothorac Surg 2014;45:408-16. [Crossref] [PubMed]
- Paget S. The distribution of secondary growths in cancer of the breast. 1889. Cancer Metastasis Rev 1989;8:98-101. [PubMed]
- Schweiger T, Hegedus B, Nikolowsky C, et al. EGFR, BRAF and KRAS status in patients undergoing pulmonary metastasectomy from primary colorectal carcinoma: a prospective follow-up study. Ann Surg Oncol 2014;21:946-54. [Crossref] [PubMed]
- Lo Iacono M, Monica V, Righi L, et al. Targeted next-generation sequencing of cancer genes in advanced stage malignant pleural mesothelioma: a retrospective study. J Thorac Oncol 2015;10:492-9. [Crossref] [PubMed]
- Konig K, Peifer M, Fassunke J, et al. Implementation of Amplicon Parallel Sequencing Leads to Improvement of Diagnosis and Therapy of Lung Cancer Patients. J Thorac Oncol 2015;10:1049-57. [Crossref] [PubMed]
- Xiao N, Xu QS, Li MZ. hdnom: Building Nomograms for Penalized Cox Models with High-Dimensional Survival Data. bioRxiv 2016;065524. [Crossref]
- Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 2017;45:D362-8. [Crossref] [PubMed]
- Tsilimigras DI, Ntanasis-Stathopoulos I, Bagante F, et al. Clinical significance and prognostic relevance of KRAS, BRAF, PI3K and TP53 genetic mutation analysis for resectable and unresectable colorectal liver metastases: A systematic review of the current evidence. Surg Oncol 2018;27:280-8. [Crossref] [PubMed]
- Tie J, Lipton L, Desai J, et al. KRAS mutation is associated with lung metastasis in patients with curatively resected colorectal cancer. Clin Cancer Res 2011;17:1122-30. [Crossref] [PubMed]
- Crumley SM, Pepper KL, Phan AT, et al. Next-Generation Sequencing of Matched Primary and Metastatic Rectal Adenocarcinomas Demonstrates Minimal Mutation Gain and Concordance to Colonic Adenocarcinomas. Arch Pathol Lab Med 2016;140:529-35. [Crossref] [PubMed]
- Crobach S, Ruano D, van Eijk R, et al. Somatic mutation profiles in primary colorectal cancers and matching ovarian metastases: Identification of driver and passenger mutations. J Pathol Clin Res 2016;2:166-74. [Crossref] [PubMed]
- Kovaleva V, Geissler AL, Lutz L, et al. Spatio-temporal mutation profiles of case-matched colorectal carcinomas and their metastases reveal unique de novo mutations in metachronous lung metastases by targeted next generation sequencing. Mol Cancer 2016;15:63. [Crossref] [PubMed]
- Vignot S, Lefebvre C, Frampton GM, et al. Comparative analysis of primary tumour and matched metastases in colorectal cancer patients: evaluation of concordance between genomic and transcriptional profiles. Eur J Cancer 2015;51:791-9. [Crossref] [PubMed]
- Renaud S, Romain B, Falcoz PE, et al. KRAS and BRAF mutations are prognostic biomarkers in patients undergoing lung metastasectomy of colorectal cancer. Br J Cancer 2015;112:720-8. [Crossref] [PubMed]
- Shindoh J, Nishioka Y, Yoshioka R, et al. KRAS Mutation Status Predicts Site-Specific Recurrence and Survival After Resection of Colorectal Liver Metastases Irrespective of Location of the Primary Lesion. Ann Surg Oncol 2016;23:1890-6. [Crossref] [PubMed]
- Ghidini M, Personeni N, Bozzarelli S, et al. KRAS mutation in lung metastases from colorectal cancer: prognostic implications. Cancer Med 2016;5:256-64. [Crossref] [PubMed]
- Pereira AA, Rego JF, Morris V, et al. Association between KRAS mutation and lung metastasis in advanced colorectal cancer. Br J Cancer 2015;112:424-8. [Crossref] [PubMed]
- Zhang YL, Wang RC, Cheng K, et al. Roles of Rap1 signaling in tumor cell migration and invasion. Cancer Biol Med 2017;14:90-9. [Crossref] [PubMed]
- Dallol A, Buhmeida A, Al-Ahwal MS, et al. Clinical significance of frequent somatic mutations detected by high-throughput targeted sequencing in archived colorectal cancer samples. J Transl Med 2016;14:118. [Crossref] [PubMed]
- Yang Y, Forslund A, Remotti H, et al. P53 mutations in primary tumors and subsequent liver metastases are related to survival in patients with colorectal carcinoma who undergo liver resection. Cancer 2001;91:727-36. [Crossref] [PubMed]
- Wehler TC, Frerichs K, Graf C, et al. PDGFRalpha/beta expression correlates with the metastatic behavior of human colorectal cancer: a possible rationale for a molecular targeting strategy. Oncol Rep 2008;19:697-704. [PubMed]
- Johnson B, Cooke L, Mahadevan D. Next generation sequencing identifies 'interactome' signatures in relapsed and refractory metastatic colorectal cancer. J Gastrointest Oncol 2017;8:20-31. [Crossref] [PubMed]
- Pancione M, Remo A, Zanella C, et al. The chromatin remodelling component SMARCB1/INI1 influences the metastatic behavior of colorectal cancer through a gene signature mapping to chromosome 22. J Transl Med 2013;11:297. [Crossref] [PubMed]
- Wang J, Andrici J, Sioson L, et al. Loss of INI1 expression in colorectal carcinoma is associated with high tumor grade, poor survival, BRAFV600E mutation, and mismatch repair deficiency. Hum Pathol 2016;55:83-90. [Crossref] [PubMed]
- Jauhri M, Bhatnagar A, Gupta S, et al. Targeted molecular profiling of rare genetic alterations in colorectal cancer using next-generation sequencing. Med Oncol 2016;33:106. [Crossref] [PubMed]
- Hedegaard J, Thorsen K, Lund MK, et al. Next-generation sequencing of RNA and DNA isolated from paired fresh-frozen and formalin-fixed paraffin-embedded samples of human cancer and normal tissue. PLoS One 2014;9:e98187. [Crossref] [PubMed]


