Association between RUNX3 promoter methylation and non-small cell lung cancer: a meta-analysis
Background
Lung cancer is one of the most common malignancy and a leading cause of cancer-related deaths worldwide (1). Non-small cell lung cancer (NSCLC) accounts for 80% of all primary lung cancers, which comprises adenocarcinoma, squamous cell carcinoma and large cell carcinoma (2). Current knowledge regarding epigenetic changes play an integral role in the transformation, promotion and progression of cancer (3,4). DNA methylation is one of the most common forms of epigenetic modification. The abnormal hypermethylation patterns of promoter site in various tumor suppressor genes (TSGs) are a pivotal mechanism in a wide variety of malignancies including lung cancer (5,6).
In humans 1p36.1 chromosomal loci, where RUNX3 is located at this locus, observed to undergo frequent deletion could induce pulmonary carcinogenesis (7,8). RUNX3 is a known regulator in the transforming growth factor (TGF)-β signaling pathway, which has recently been reported as a candidate tumor suppressor (9-11). Decreased RUNX3 expression or deletion are mainly due to methylation or allelic loss, that results in the limited function of Smad proteins and the promotion of TGF-β signaling, which leads to tumor development (12). Previous studies have demonstrated RUNX3 promoter methylation playing a crucial role in neoplasias, including colorectal (13), gastric (14,15), lung (16), bladder (17), breast (18,19) oral (20), and liver cancers (21), either using cell lines, or primary cancer tissues. However, the relationship between RUNX3 promoter methylation and NSCLC remains to be clarified.
Although this association has been investigated in separate studies, the results are somewhat contradictory (22,23), possibly due to small sample size and underpowered in a single study. Therefore, we performed a meta-analysis using all available related studies to assess the association of RUNX3 promoter methylation and NSCLC.
Methods
Search strategies and selection criteria
We searched Pubmed, Embase, Cochrane Central, and Chinese Biological Medicine database, for articles published in English or Chinese. We identified the publications using the text words (RUNX3 or PEBP2αC or CBFA3 or AML2) AND (lung or pulmonic) AND (cancer or neoplasm). The search updated on March 7, 2014. In addition, we also reviewed the reference from retrieved papers and relevant review articles. We only recruited data from fully published papers, not meeting or conference abstracts.
Study selection
Two investigators (Yali Liang and Lianping He) first independently screened the titles and abstracts to identify relevant articles. A second screening was based on full-text articles to further see whether they had met the inclusion criteria. Discrepancies were resolved by consensus.
Studies were included if they meet the following criteria: (I) the specimens from peripheral serum or surgically respected primary tumor tissue (not cell line or sputum); (II) the exposure of interest was RUNX3 promoter methylation; (III) the outcome of interest was NSCLC; (IV) odds ratio (OR) with corresponding 95% confidence intervals (CIs) (or data to calculate them) were published.
Quality assessment
Two investigators independently assessed methodological quality of eligible studies with the Newcastle-Ottawa scale (NOS). The quality scale consists of three parameters: selection, comparability, and exposure assessment. The quality score ranges from 0 to 9. Studies with a score equal to or higher than 4 were considered “high-quality”, whereas those scored less than 4 were considered “low-quality”.
Data extraction
In order to control the bias and improve the reliability, the investigator followed a standardized data-collection form to extract all data. The following information was collected from each study: first author, year of publication, country of the study objects, specimen origin, number of cases and controls, the methylation status of RUNX3 promoter in cancer and control samples, correlation between methylation and clinicopathological characteristics in NSCLC.
Statistical analysis
Our main analyses were focused on the association between RUNX3 promoter methylation and risk of NSCLC. The effect measures of interest were ORs and corresponding 95% CI for case-control study.
Heterogeneity test for pooled ORs was performed by I2 statistic (statistically significant level at I2 ≥50%) (24). If heterogeneity was observed, we used random effects model (DerSimonian-Laird method) to calculate the overall odds ratios, otherwise fixed effects model (Mantel-Haenszel) was used. Subgroup analysis was performed according to specimen types (lung tissue or peripheral serum), and age categories (≤60 years; between 60 and 65 years; >65 years). If the heterogeneity was strong, meta-regression analyses were employed to analyze the sources of the heterogeneity. Sensitivity analysis was performed by deleting one study in each turn to evaluate the stability of the final results. A funnel plot and Egger’s test were conducted to investigate any potential publication bias. We also assessed the correlation between methylation status and clinicopathological characteristics (gender, smoking history, tumor stage, differentiation and histopathology) in NSCLC; histopathological tumor type includes adenocarcinoma, squamous cell carcinoma, and other histological type (large cell carcinoma and mixed histologies carcinoma).
The statistical analyses were performed with Stata12.0 software and review manager 4.2, two-sided P values less than 0.05 were considered statistically significant.
Results
Study selection and characteristics
One hundred and four potentially relevant studies were identified by the electronic search strategy, and 1 study was obtained by manually searching all references cited in the original studies. We justified eligible studies by further screening of their titles, abstracts and full texts. As a result, we retrieved 21 potentially relevant articles. Eight studies were excluded, because one studies did not exactly define as NSCLC (25); one study measured RUNX3 methylation status by the RT-PCR (22); and one study data there had errors (26); and four studies did not establish control groups (27-30). Finally, the remaining 13 studies included in our study (Figure 1) (23,31-42) were included in our meta-analysis. All of them were case-control studies, and two papers were written in Chinese. The main characteristics of the reviewed studies are showed in Table 1. The total sample size was 1,368 (759 cases and 609 controls).

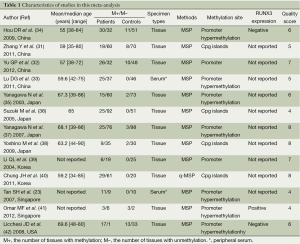
Full table
RUNX3 promoter methylation and risk of NSCLC
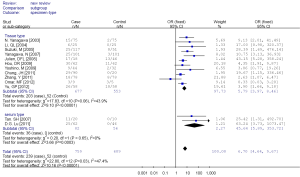
Among these 13 studies, substantial heterogeneity was not obvious (I2 =47.4%). Hence, fixed effects model (Mantel-Haenszel) was used to calculate the pooled OR and 95% CIs (Figure 2). Overall, the pooled OR for RUNX3 methylation in cancer specimens compared with normal specimens was 6.70 (95% CI: 4.64-9.67).
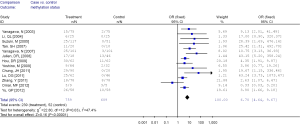
Subgroup analysis and meta-regression
Subgroup analysis was performed according to specimen types (tissue or peripheral serum) and age categories (≤60 years; between 60 and 65 years; >65 years). In the analysis of specimen-types subgroup, the summary OR was 5.79 (95% CI: 3.97-8.46) for tissue specimen subgroup, and that was 45.64 (95% CI: 5.89-353.72) for serum specimen subgroup. There was no evidences of heterogeneity in different specimen types subgroup (I2 =43.9%; I2 =0%). The OR for the age ≤60 years subgroup was 5.19 (95% CI: 3.27-8.24), that for the 60-65 years group was 9.45 (95% CI: 2.45-36.45) and that for the >65 years subgroup was 13.23 (95% CI: 5.59-31.28). Heterogeneity was not observed within different age categories subgroup (I2 =39.8%; I2 =43.8%; I2 =0%), as showed in Figures 3 and 4.
Heterogeneity was borderline among these 13 studies (I2 =47.4%). Therefore, we performed further analyses to detect the sources of the heterogeneity using the meta-regression method with restricted maximum likelihood modification. The result of meta-regression indicated that the trend in ORs was correlated with age, which accounted for the heterogeneity (coefficient =0.61, P=0.046, adjusted R2 =100%). However, the other factor (specimen type) could not explain the heterogeneity. The results are showed in Table 2.

Full table
Sensitivity analysis and publication bias
We conducted sensitivity analysis to assess the stability of the overall effects by deleting a single study, the overall ORs did not substantially changed, with a range from 6.00 (95% CI: 4.13-8.73) to 7.84 (95% CI: 5.21-11.79).
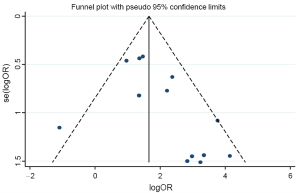
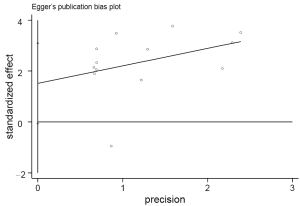
The funnel plot and Egger’s test were performed to estimate publication bias (Figures 5,6), there was no evidence of publication bias with regard to RUNX3 methylation in relation to NSCLC risk (Egger’s test: t=2.12, P=0.058).
Correlation between methylation of RUNX3 promoter and clinicopathological characters
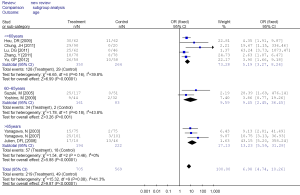
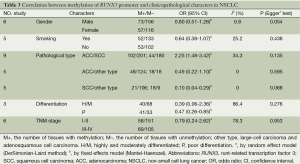
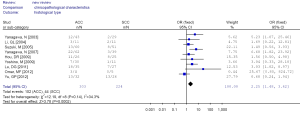
Table 3 showed the correlation between methylation of RUNX3 promoter and clinicopathological characters. A portion of these 13 studies provided the relationship of RUNX3 methylation and clinicopathological characteristics in their cancer specimens. The overall results demonstrated that RUNX3 methylation was less frequent in adenocarcinoma compared with squamous cell carcinoma (OR =2.25, 95% CI: 1.48-3.42), but the frequencies was similar in adenocarcinoma and other histological type (OR =0.49, 95% CI: 0.22-1.10). Highly and moderately differentiated cancer specimens also had a lower methylation than poor differentiation (OR =0.39, 95% CI: 0.06-2.36).
Full table
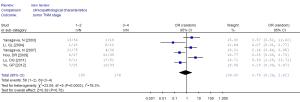
There were no significant differences in RUNX3 methylation status in cancer sample in relation to gender, smoking and tumor stage. The results are showed in Figures 7-12.


Discussion
Our meta-analysis focuses on relationship between RUNX3 promoter methylation and the risk of NSCLC. The overall OR for methylation status in cancer versus normal specimens was 6.70 (95% CI: 4.64-9.67) by a fixed effects model on pooled data from 13 studies. In the specimen types-specific subgroup analysis, results showed: the OR in the tissue sample subgroup was 5.79 (95% CI: 3.97-8.46), that in serum samples subgroup was 45.64 (95% CI: 5.89-353.72), which further confirmed RUNX3 methylation was a potential risk factor for NSCLC. Among peripheral blood of non-cancer objects, the methylation of RUNX3 was absent, and so the methylation of RUNX3 could be regarded as cancer-specific phenomenon. Therefore, in terms of clinical application, the detection RUNX3 methylation of peripheral blood may be useful as a diagnostic approach. The trend of association between RUNX3 methylation and NSCLC was correlated with age. The OR was 5.19 (95% CI: 3.27-8.24) for the age ≤60 years subgroup, 9.45 (95% CI: 2.45-36.45) for the group of 60-65 years, and 13.23 (95% CI: 5.59-31.28) for the >65 years subgroup. The coefficient for age was calculated to be 0.61 by meta-regression analysis, indicating that the tendency for RUNX3 methylation increased with age. DNA methylation, genomic imprinting, and histone modifications were examples of epigenetic factors known to undergo change in the aging and malignant counterparts (43). RUNX3 exhibited altered DNA methylation patterns in aging, displaying sometimes tissue- and cell type-specific features with consequent different functional outcomes (44). Some studies reported RUNX3 methylation occurring preferentially in older malignant tumor patients (45). These results suggested that RUNX3 methylation related with individual age in malignancies. We found that the ORs for RUNX3 methylation increased from 5.19 in the younger age group, through 9.45, to 13.33 in the oldest age group. The coefficient for age was calculated to be 0.61 by meta-regression analysis, indicating that the tendency for RUNX3 methylation increased with advancing age. The results suggested RUNX3 methylation may be preferentially occurs in older NSCLC patients.
Although no publication bias was detected using Egger’s test, the funnel plot showed one of the studies exceeded the 95% confidence limits. We performed sensitivity analysis in each to estimate the robustness of our results by deleting one study. The overall ORs were slightly changed from 6.00 (95% CI: 4.13-8.73) to 7.84 (95% CI: 5.21-11.79), which were still significant. To produce a more robust estimation, we performed sensitivity analysis by deleting one study in each turn. The overall ORs were slightly changed from 6.00 (95% CI: 4.13-8.73) to 7.84 (95% CI: 5.21-11.79), which were still significant, indicating a strong association between RUNX3 promoter methylation and NSCLC.
There were no significant differences in RUNX3 methylation in cancer tissues in relation to gender, smoking history, or tumor TNM stage.
The aggregated results found that RUNX3 methylation was less frequent in squamous cell carcinoma compared with adenocarcinoma and other histological type, suggesting that inactivation of RUNX3 might play a less significant role in the pathogenesis of squamous cell carcinoma, as those previous studies suggested (30).
Analysis of the pooled data also showed that undifferentiated NSCLC had a higher frequency of promoter methylation than well-differentiated, which was significant in the fixed model (OR =0.47, CI: 0.26-0.85) but non-significant in the random effects model. This phenomenon may be associated with the smaller number of studies analyzed. But the results also indicated that RUNX3 promoter methylation may be related to poor prognosis.
Some limitations of this meta-analysis should be addressed. First, Although we searched literature as completely as possible, the results calculated in our meta-analysis maybe exist bias as we only collected full published papers and articles published in English or Chinese. Second, our results were based on unadjusted, whereas a more precise analysis should be conducted if adjustment estimates were available.
Conclusions
Despite these limitations, this meta-analysis provides additional evidence to support a strong association between methylation of the RUNX3 promoter and NSCLC. The tendency of association for RUNX3 methylation increased with age. The RUNX3 methylation was also associated with histological type of the NSCLC. However, it is necessary to conduct large sample studies using standardized and well-matched controls.
Acknowledgements
All authors have made substantial contributions to this article: Yingshui Yao contributed to the conception and design. Yali Liang contributed to the analysis and interpretation of data, and drafting of the article. Lianping He contributed to the acquisition of data and revision of the article. Hui Yuan and Yuelong Jin contributed to discussion of the article design. All authors read and approved the final manuscript.
Disclosure: The authors declare no conflict of interest.
References
- Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2009. CA Cancer J Clin 2009;59:225-49. [PubMed]
- Stinchcombe TE, Socinski MA. Current treatments for advanced stage non-small cell lung cancer. Proc Am Thorac Soc 2009;6:233-41. [PubMed]
- Esteller M, Corn PG, Baylin SB, et al. A gene hypermethylation profile of human cancer. Cancer Res 2001;61:3225-9. [PubMed]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002;3:415-28. [PubMed]
- Risch A, Plass C. Lung cancer epigenetics and genetics. Int J Cancer 2008;123:1-7. [PubMed]
- Shames DS, Girard L, Gao B. A genome-wide screen for promoter methylation in lung cancer identifies novel methylation markers for multiple malignancies. PLoS Med 2006;3:e486. [PubMed]
- Herzog CR, Wang Y, You M. Allelic loss of distal chromosome 4 in mouse lung tumors localize a putative tumor suppressor gene to a region homologous with human chromosome 1p36. Oncogene 1995;11:1811-5. [PubMed]
- Nomoto S, Haruki N, Tatematsu Y, et al. Frequent allelic imbalance suggests involvement of a tumor suppressor gene at 1p36 in the pathogenesis of human lung cancers. Genes Chromosomes Cancer 2000;28:342-6. [PubMed]
- Hanai J, Chen LF, Kanno T, et al. Interaction and functional cooperation of PEBP2/CBF with Smads. Synergistic induction of the immunoglobulin germline Calpha promoter. J Biol Chem 1999;274:31577-82. [PubMed]
- Lee JM, Kwon HJ, Bae SC, et al. Lung tissue regeneration after induced injury in Runx3 KO mice. Cell Tissue Res 2010;341:465-70. [PubMed]
- Lee KS, Lee YS, Lee JM, et al. Runx3 is required for the differentiation of lung epithelial cells and suppression of lung cancer. Oncogene 2010;29:3349-61. [PubMed]
- Bae SC, Choi JK. Tumor suppressor activity of RUNX3. Oncogene 2004;23:4336-40. [PubMed]
- Subramaniam MM, Chan JY, Soong R, et al. RUNX3 inactivation in colorectal polyps arising through different pathways of colonic carcinogenesis. Am J Gastroenterol 2009;104:426-36. [PubMed]
- Chen W, Gao N, Shen Y, et al. Hypermethylation downregulates Runx3 gene expression and its restoration suppresses gastric epithelial cell growth by inducing p27 and caspase3 in human gastric cancer. J Gastroenterol Hepatol 2010;25:823-31. [PubMed]
- Homma N, Tamura G, Honda T, et al. Spreading of methylation within RUNX3 CpG island in gastric cancer. Cancer Sci 2006;97:51-6. [PubMed]
- Yanada M, Yaoi T, Shimada J, et al. Frequent hemizygous deletion at 1p36 and hypermethylation downregulate RUNX3 expression in human lung cancer cell lines. Oncol Rep 2005;14:817-22. [PubMed]
- Jeong P, Min BD, Ha YS, et al. RUNX3 methylation in normal surrounding urothelium of patients with non-muscle-invasive bladder cancer: potential role in the prediction of tumor progression. Eur J Surg Oncol 2012;38:1095-100. [PubMed]
- Lau QC, Raja E, Salto-Tellez M, et al. RUNX3 is frequently inactivated by dual mechanisms of protein mislocalization and promoter hypermethylation in breast cancer. Cancer Res 2006;66:6512-20. [PubMed]
- Subramaniam MM, Chan JY, Soong R, et al. RUNX3 inactivation by frequent promoter hypermethylation and protein mislocalization constitute an early event in breast cancer progression. Breast Cancer Res Treat 2009;113:113-21. [PubMed]
- de Freitas Cordeiro-Silva M, Stur E, Agostini LP, et al. Promoter hypermethylation in primary squamous cell carcinoma of the oral cavity and oropharynx: a study of a Brazilian cohort. Mol Biol Rep 2012;39:10111-9. [PubMed]
- Park WS, Cho YG, Kim CJ, et al. Hypermethylation of the RUNX3 gene in hepatocellular carcinoma. Exp Mol Med 2005;37:276-81. [PubMed]
- Jin M, Kawakami K, Fukui Y, et al. Different histological types of non-small cell lung cancer have distinct folate and DNA methylation levels. Cancer Sci 2009;100:2325-30. [PubMed]
- Tan SH, Ida H, Lau QC, et al. Detection of promoter hypermethylation in serum samples of cancer patients by methylation-specific polymerase chain reaction for tumour suppressor genes including RUNX3. Oncol Rep 2007;18:1225-30. [PubMed]
- Labeau SO, Van de Vyver K, Brusselaers N, et al. Prevention of ventilator-associated pneumonia with oral antiseptics: a systematic review and meta-analysis. Lancet Infect Dis 2011;11:845-54. [PubMed]
- Kim TY, Lee HJ, Hwang KS, et al. Methylation of RUNX3 in various types of human cancers and premalignant stages of gastric carcinoma. Lab Invest 2004;84:479-84. [PubMed]
- Castro M, Grau L, Puerta P, et al. Multiplexed methylation profiles of tumor suppressor genes and clinical outcome in lung cancer. J Transl Med 2010;8:86. [PubMed]
- Suzuki M, Wada H, Yoshino M, et al. Molecular characterization of chronic obstructive pulmonary disease-related non-small cell lung cancer through aberrant methylation and alterations of EGFR signaling. Ann Surg Oncol 2010;17:878-88. [PubMed]
- Yanagawa N, Tamura G, Oizumi H, et al. Inverse correlation between EGFR mutation and FHIT, RASSF1A and RUNX3 methylation in lung adenocarcinoma: relation with smoking status. Anticancer Res 2011;31:1211-4. [PubMed]
- Tang Y, Wu F, Hu C. RUNX3 promoter hypermethylation and prognosis of early surgically resected non-small cell lung cancers. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2011;36:650-4. [PubMed]
- Sato K, Tomizawa Y, Iijima H, et al. Epigenetic inactivation of the RUNX3 gene in lung cancer. Oncol Rep 2006;15:129-35. [PubMed]
- Zhang Y, Wang R, Song H, et al. Methylation of multiple genes as a candidate biomarker in non-small cell lung cancer. Cancer Lett 2011;303:21-8. [PubMed]
- Yu GP, Ji Y, Chen GQ, et al. Application of RUNX3 gene promoter methylation in the diagnosis of non-small cell lung cancer. Oncol Lett 2012;3:159-162. [PubMed]
- Lu DG, Ji XQ, Liu W. Significance of RUNX3 Hypermethylation in Serum DNA of Non-small Cell Lung Cancer Patients. Cancer Research on Prevention and Treatment 2011;38:671-4.
- Hou DR, Wang HZ. Analysis of promoter hypermethylation of Runx3 gene in non-small cell lung carcinoma. Progress in Modern Biomedicine 2009;9:3692-5.
- Yanagawa N, Tamura G, Oizumi H, et al. Promoter hypermethylation of tumor suppressor and tumor-related genes in non-small cell lung cancers. Cancer Sci 2003;94:589-92. [PubMed]
- Suzuki M, Shigematsu H, Shames DS, et al. DNA methylation-associated inactivation of TGFbeta-related genes DRM/Gremlin, RUNX3, and HPP1 in human cancers. Br J Cancer 2005;93:1029-37. [PubMed]
- Yanagawa N, Tamura G, Oizumi H, et al. Promoter hypermethylation of RASSF1A and RUNX3 genes as an independent prognostic prediction marker in surgically resected non-small cell lung cancers. Lung Cancer 2007;58:131-8. [PubMed]
- Yoshino M, Suzuki M, Tian L, et al. Promoter hypermethylation of the p16 and Wif-1 genes as an independent prognostic marker in stage IA non-small cell lung cancers. Int J Oncol 2009;35:1201-9. [PubMed]
- Li QL, Kim HR, Kim WJ, et al. Transcriptional silencing of the RUNX3 gene by CpG hypermethylation is associated with lung cancer. Biochem Biophys Res Commun 2004;314:223-8. [PubMed]
- Chung JH, Lee HJ, Kim BH, et al. DNA methylation profile during multistage progression of pulmonary adenocarcinomas. Virchows Arch 2011;459:201-11. [PubMed]
- Omar MF, Ito K, Nga ME, et al. RUNX3 downregulation in human lung adenocarcinoma is independent of p53, EGFR or KRAS status. Pathol Oncol Res 2012;18:783-92. [PubMed]
- Licchesi JD, Westra WH, Hooker CM, et al. Epigenetic alteration of Wnt pathway antagonists in progressive glandular neoplasia of the lung. Carcinogenesis 2008;29:895-904. [PubMed]
- Damaschke NA, Yang B, Bhusari S, et al. Epigenetic susceptibility factors for prostate cancer with aging. Prostate 2013;73:1721-30. [PubMed]
- D’Aquila P, Rose G, Bellizzi D, et al. Epigenetics and aging. Maturitas 2013;74:130-6. [PubMed]
- Waki T, Tamura G, Sato M, et al. Age-related methylation of tumor suppressor and tumor-related genes: an analysis of autopsy samples. Oncogene 2003;22:4128-33. [PubMed]