
Complicated prognostic values of CCL28 in breast cancer by subtype
Introduction
Breast cancer is the most prevalent malignancy and the leading cause of cancer death among women worldwide (1). As a heterogeneous collection of diseases, breast cancer is molecularly classified into different subtypes, leading to various prognostic evaluation and therapeutic decision making (2). An increasing number of studies have focused on the novel biomarkers so as to provide better and comprehensive prognostic evaluation for breast cancer patients along with the current practical prognostic factors [e.g., estrogen receptor (ER), progesterone receptor (PR), HER-2 and Ki-67] (3,4). However, it remains unclear that whether the same biomarker is able to assess different subtypes of cancer consistently or at least in the same direction.
Chemokines are small cytokine-like secreted proteins primarily secreted by stromal cells with chemo-attractant properties (5-7). Previous findings have uncovered the significance of chemokines in cancer biology, by not only mediating immune-cells homing into tumors but also being directly involved in the regulation of the development, progression, invasion and metastasis of different human malignancies (8,9). Our previous research also revealed the involvement of some chemokines in breast cancer. For example, CCR4 was shown to promote tumor growth and lung metastasis in breast cancer (10). CXCL14 was found to play an inhibitory role in the proliferation and metastasis of breast malignancy (11).
One of the members of CC subfamily, CCL28 or mucosae-associated epithelial chemokine, is a ligand for CCR3/CCR10 and typically secreted from epithelial cells in the gut, lung salivary, as well as breast gland (12,13). Several studies have shown its functions on mucosal immunity and antimicrobial activity (14). This chemokine also plays divergent roles in a variety of cancers. Facciabene et al. have discovered for the first time that the overexpression of CCL28 under hypoxia could account for the shortened survival in patients with ovarian cancer (15). On the contrary, another clinical research found the notably lower expression of the CCL28 protein in colorectal tumors compared with normal tissues (16). The expression of CCL28, as well as its relationship with tumor biological behaviors and patients’ survival outcomes, remain unclear in the setting of heterogeneous breast cancer. One study found that CCL28 mRNA expression was eliminated in human breast tumors compared to adjacent normal tissues (17). However, Yang et al. demonstrated that highly expressed CCL28 served as a detrimental factor through promoting the proliferation, invasiveness and metastasis of MDA-MB-231 breast cancer cell line (18). Taken together, the prognostic relevance of CCL28 in different subtypes of breast cancer requires further exploration. The current study was designed to investigate the prognostic values of CCL28 in breast cancer with a focus on divergent subtypes. Extended validation was performed in the cohorts from publicly available datasets.
Methods
Study patients and samples
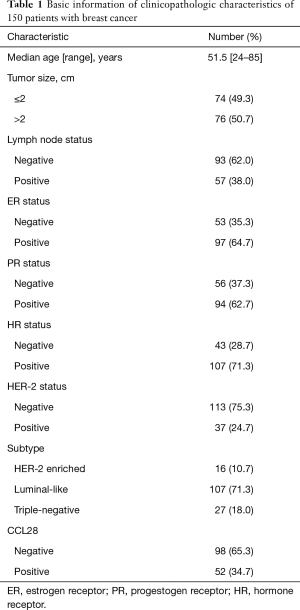
We included 150 patients in the discovery cohort, with ages ranging from 24 to 85 years and the median follow-up time for 50 months. The basic information of clinicopathologic features is shown in Table 1. All samples were collected between May-2010 and March-2013, at the Department of Breast Surgery in Affiliated Cixi Hospital of Wenzhou Medical University. We staged the participants according to the American Joint Committee on Cancer (AJCC) pathologic tumor-node-metastasis (TNM) classification (the 7th edition). All patients included were systematically untreated before the surgery, with operable primary invasive breast carcinoma (stages I to IIb and T3N1M0). Those with neoadjuvant chemotherapy, prior malignancies, and stage IV diseases were excluded. Our research protocol had been approved by the Ethical Committee of the Affiliated Cixi Hospital of Wenzhou Medical University, with all participants providing written informed consents.
Full table
Immunohistochemistry (IHC) assay for CCL28 in the discovery cohort
We performed IHC assay for tumorous expression of CCL28 in the discovery cohort. The procedure of IHC assay had been described previously (19). With four-micron paraffin sections prepared, tissue sections were deparaffinized in xylene for 5 minutes and rehydrated with graded ethanol. Three percent hydrogen peroxide was used to inhibit the endogenous peroxidase activity, and antigen retrieval was conducted using 10 mM citrate buffer. Afterward, the sections were incubated with diluted goat serum for 10 minutes followed by overnight incubation with primary CCL28 rabbit monoclonal antibody (dilution 1:500, rabbit polyclonal to CCL28, clone PA5-28821, Thermo Corp. USA) at 4 °C. After incubation with biotinylated secondary antibody for 30 minutes and with streptavidin-peroxidase for 30 minutes, staining development was finally conducted with 3-3'-diaminobenzidine. Phosphate buffer saline, instead of primary antibody, was used for negative controls.
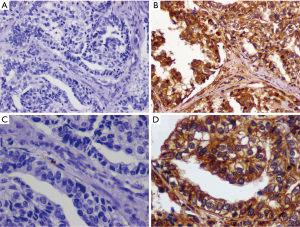
Immune-reactivity was estimated according to Hao’s method (20). We evaluated the cytoplasmic and membranaceous staining intensity and percentage of positive tumor cells in five randomly selected high-power fields (200×) for each sample. The extent of staining was divided into five semi-quantitative categories based on the percentages of positive tumor cells: 0 (<5% positive cells), 1 (6% to 25% positive cells), 2 (26% to 50% positive cells), 3 (51% to 75% positive cells) and 4 (>75% positive cells). In addition, the intensity of staining was classified semi-quantitatively from 0 to 3: 0 for negative, 1 for weak staining, 2 for moderate staining and 3 for strong staining. The intensity and percentage scores were multiplied to obtain the final staining scores: 0 to 2, as negative; 3 to 12, as positive. All specimens were evaluated independently by two pathologists who were unaware of patients’ clinicopathologic information. In the cases of score discrepancies, the slides were re-examined, and two observers reached consensuses through discussion. The representative CCL28 IHC staining plots are shown in Figure 1.
Measurement of ER, PR, and HER-2 status in the discovery cohort
We conducted IHC analysis of ER, PR, and HER-2 expression in the paraffin-embedded tumor surgical specimens of our participants, according to the guideline of American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP) (21,22). At least 1% tumor nuclei immunoreactive for ER or PR should be considered positive (21). HER-2 was determined positive by IHC 3+ (HerceptTest, DAKO, Denmark) or fluorescence in situ hybridization (FISH) positive status (PathVysion HER-2 DNA probe kit) (22). ER and PR were merged into hormone receptor (HR) with HR status positive for ER and/or PR positive, HR status negative for ER and PR negative. Then we categorized the cases in our cohort into three clinical subtypes based on the HR and HER-2 status: HER-2 enriched (HR negative, HER-2 positive), luminal-like (HR positive, HER-2 negative or positive), triple-negative (HR and HER-2 negative). Consequently, there were 16 (10.7%) HER-2 enriched, 107 (71.3%) luminal-like and 27 (18.0%) triple-negative cases.
Validation cohorts
Extended validation was carried out in two publicly available databases. The first cohort was from the Cancer Genome Atlas (TCGA) database. The data on clinical information were downloaded from Cbioportal (http://www.cbioportal.org/) on October 07, 2017 (23,24). The data on CCL28 expression were extracted from FPKM-UQ files (CCL28/ENSG00000151882.10) downloaded from the GDC data portal (https://portal.gdc.cancer.gov) on October 27, 2017 (25,26). Only cases staging from I to III, with available data on HR status, HER-2 status, tumor size, lymph node status, CCL28 mRNA expression (RNA sequencing) and disease-free survival (DFS) were included, resulting in a cohort of 863 patients. To consist with the grouping of the discovery cohort, the patients in the TCGA cohort was categorized into the same three clinical subtypes according to the HR and HER-2 status measured by IHC and FISH assay: HER-2 enriched, luminal-like and triple-negative breast cancer, with the cases number of 35, 683 and 145, respectively. The median expression of CCL28 in these 863 patients was decided as the cutoff value of high and low expression subgroups.
Another online database named Kaplan-Meier plotter was established with data on the gene expression and survival information of breast cancer patients downloaded from Gene Expression Omnibus, European Genome-Phenome Archive and TCGA (27,28), last updated in October 13, 2016 and containing now 5,143 samples. A total of 35 datasets of breast cancer were utilized for analysis, with a detailed list available on the webpage (http://kmplot.com/analysis/index.php?p=service&cancer=breast). Redundant samples (repeated microarrays representing the same individuals) and biased arrays (two or more parameters are out of 95% range of all arrays) were removed for quality control, with restriction on follow-up time of no more than 120 months, leading to 1,764 breast cancer cases with data on the mRNA expression of desired gene CCL28 (probe ID, 238750_at) and corresponding relapse-free survival (RFS) information. Subgroup analysis meeting the purpose of our research was based on the 2013 St. Gallen molecular classification criteria using the expression of ER-1 gene, HER-2 gene, and marker of proliferation Ki-67 gene (29). Among 1,764 cases with breast cancer, the number of luminal A, luminal B, triple-negative phenotype and HER-2 enriched cases were 841, 407, 360 and 156, respectively. Patients with high or low CCL28 expression were grouped using “Auto select best cutoff” (calculate all percentiles and automatically select the best execution threshold as the cutoff value).
Statistical analysis
In the discovery and Kaplan-Meier cohorts, the study endpoint of interest was RFS, defined as the time from initial diagnosis biopsy to the occurrence of local or regional recurrence, or distance metastasis. In the TCGA cohort, the primary outcome was DFS, which was calculated from initial diagnosis biopsy to local or regional recurrence, distance metastasis, or death as a result of any non-breast cancer cause. Given that the main endpoint events of DFS were recurrence and metastasis, RFS and DFS could be considered similar when appropriate. The maximum follow-up time was restricted to 120 months. The χ2 test or Fisher’s exact test was applied to estimate the association of CCL28 expression with clinicopathologic variables. The effects of CCL28 expression on survival outcomes in patients from the discovery cohort and TCGA cohort were evaluated by univariate Cox regression analysis as well as multivariate Cox regression analysis adjusted by tumor size, lymph node status, HR status and HER-2 status. Kaplan-Meier method and log-rank test were applied to plot and compare the survival curves. As for the Kaplan-Meier potter cohort, survival analysis was performed online, and survival curves were plotted on the webpage with the calculated hazard ratio (HR), 95% confidence intervals (CI), and log-rank P values. All of these statistical analyses were performed using SPSS 23.0 (IBM Corp, Armonk, NY). Statistically significant was defined as two-sided P<0.05.
Results
Association of CCL28 expression with clinicopathologic variables
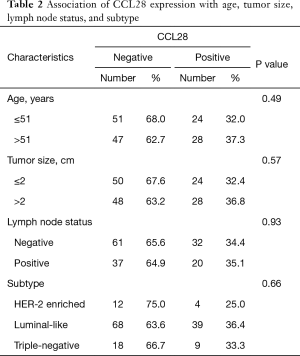
In the discovery cohort, we evaluated the relevance of CCL28 expression with clinicopathologic characteristics including age, tumor size, lymph node status, and subtype. Among 150 patients, no significant association was found between the CCL28 expression and clinicopathologic features mentioned above (P>0.05, Table 2).
Full table
The effects of CCL28 on survival outcomes
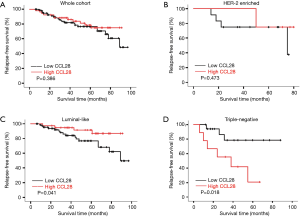
As shown in Table 3, the univariate Cox regression analysis revealed that CCL28 was not an indicator of RFS in the whole cohort (HR =0.728, 95% CI: 0.354–1.497, P=0.389) (Figure 2A) and HER-2 enriched subgroup (HR =0.451, 95% CI: 0.049–4.189, P=0.484) (Figure 2B). Interestingly, CCL28 was found related to improved RFS in luminal-like cases (HR =0.336, 95% CI: 0.112–1.008, P=0.052) (Figure 2C) but impaired RFS in patients with triple-negative breast cancer (HR =4.604, 95% CI: 1.149–18.446, P=0.031) (Figure 2D). After adjustment with tumor size and lymph node status, the results still indicated that CCL28 had no effect on RFS in the all (HR =0.770, 95% CI: 0.374–1.587, P=0.479) and HER-2 enriched (HR =0.333, 95% CI: 0.034–3.254, P=0.345) patients. Borderline statistically significant association of CCL28 with favorable RFS in luminal-like subtype (HR =0.336, 95% CI: 0.112–1.011, P=0.052) was found, in contrast to markedly shortened RFS in triple negative patients with high expression of CCL28 (HR =5.800, 95% CI: 1.342–25.074, P=0.019), which was consistent with previous univariate analysis results.

Full table
Validation for prognostic values of CCL28 in the TCGA cohort
According to the tissue sample requirements of TCGA, high-quality tumor samples with at least 80% tumor cell nuclei or powerful sequencing methods that can distinguish tumor cell signals from other cell signals are needed (30). Thus, the mRNA expression data estimated by RNA-sequencing assay from TCGA mainly revealed the presence and quantity of CCL28 mRNA inside the tumors, which was relatively consistent with the IHC assay for tumorous CCL28. Therefore, we collected data on the mRNA expression of CCL28 and DFS from TCGA dataset to validate the prognostic effects of CCL28.
In this cohort, no significant relevance of CCL28 expression with DFS was observed in the entire cohort (HR =0.943, 95% CI: 0.596–1.494, P=0.803) (Figure 3A) and HER-2 enriched subtype (HR =2.297, 95% CI: 0.207–25.460, P=0.498) (Figure 3B). In luminal-like breast cancer, favorable DFS was observed in patients displaying the high expression of CCL28 (HR =0.574, 95% CI: 0.319–1.033, P=0.064) (Figure 3C), whereas improved DFS was associated with the low CCL28 expression in triple-negative cases (HR =2.556, 95% CI: 1.059–6.169, P=0.037) (Figure 3D). After further multivariate Cox regression analysis adjusted by tumor size and lymph node status, there was also no evidence of the prognostic effect of CCL28 in the whole cohort (HR =1.028, 95% CI: 0.644–1.641, P=0.908) and HER-2 enriched subtype (HR =1.808, 95% CI: 0.163–20.079, P=0.630). CCL28 remained an independent prognostic factor for lower recurrence in luminal-like subtype (HR =0.543, 95% CI: 0.300–0.980, P=0.043) and worse DFS in patients with triple negative breast cancer (HR =3.351, 95% CI: 1.332–8.428, P=0.010) (Table 4).


Full table
Validation for prognostic effects of CCL28 in the Kaplan-Meier plotter cohort
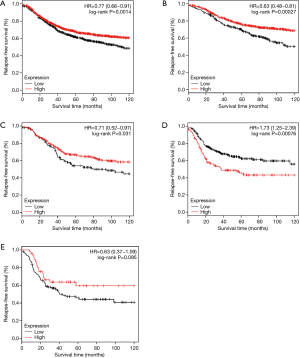
The prognostic values of CCL28 were further validated at the mRNA level in the cases from the Kaplan-Meier plotter dataset. Since multivariate analysis adjusted by tumor size and lymph node status could not be performed in this online tool, only the results of univariate analysis were shown here. Among the whole population, the high mRNA expression of CCL28 was relevant to improved RFS (HR =0.77, 95% CI: 0.66–0.91, P=0.0014) (Figure 4A). When analyzed by intrinsic subtype, CCL28 was a favorable prognostic biomarker for luminal-A (HR =0.63, 95% CI: 0.48–0.81, P<0.001) (Figure 4B), luminal-B (HR =0.71, 95% CI: 0.52–0.97, P=0.031) (Figure 4C) patients, but a poor prognostic indicator for patients with triple-negative phenotype (HR =1.73, 95% CI: 1.25–2.39, P<0.001) (Figure 4D). There was no evidence showing the significant association between CCL28 expression and RFS in HER-2 enriched participants (HR =0.63, 95% CI: 0.37–1.09, P=0.095) (Figure 4E).
Discussion
In the present study, the relevance between the CCL28 expression and survival outcomes of the patients with different breast cancer subtypes was reported. In the discovery cohort, we observed that patients with high expression of CCL28 tend to have less recurrences in luminal-like cases, but worse survival outcomes in triple-negative subtype. The observed prognostic effects of CCL28 remained after adjusted by tumor size and lymph node status. These findings were further verified in two validation cohorts from TCGA and Kaplan-Meier plotter datasets, respectively.
In addition to acting as mediators of inflammatory responses, the complicated roles of chemokines in promoting or suppressing tumor progression and metastasis has also been revealed in recent studies (31,32). Some chemokines were found to be correlated with clinical outcomes of the patients with breast cancer. For instance, chemokine CCL5 was found as a potential prognostic factor for disease progression in stage II breast cancer patients and CX3CL1 expression was associated with poor outcome in breast cancer patients (4,33). CCL28, a CC subfamily chemokine, is mainly produced by mucosal epithelial cells and involved in the development of different human malignancies, including breast cancer, with various functions (13,16,18,34). In view of the divergent prognostic roles of CCL28 in different malignancies, as well as the heterogeneity of breast cancer, the relationship between CCL28 and different subtypes of breast cancer is worth exploring.
We examined immunohistochemical CCL28 expression in 150 patients and conducted survival analysis of CCL28 on clinical outcomes. Despite no statistically significant result observed in the whole group and HER-2 enriched subgroup, it was somewhat surprising that the prognostic effects of CCL28 were complicated in breast cancer. In the luminal-like subtype, CCL28 served as a favorable predictive factor for survival, which suggests the involvement of CCL28 in the inhibitory pathway of the recurrence and metastasis of breast cancer. Contrarily, higher expression of CCL28 was correlated with poorer survival in triple-negative cases, indicating that CCL28 may act as a pro-tumor substance in this subgroup. The result obtained from triple-negative subtype was in accordance with a previous study that the overexpression of CCL28 in MDA-MB-231HM cell line resulted in enhanced tumor proliferation and metastasis via mitogen-activated protein kinase mediated, anti-apoptotic protein Bcl-2 and suppressing cell adhesion protein β-catenin involved signal pathways (18). The observed complex prognostic effects of CCL28 in different breast cancer subtypes might be attributed to the various pro-tumor or anti-tumor signaling pathways where CCL28 involved, which could be supported by several studies on the other types of malignancies. Facciabene et al. discovered that up-regulation of CCL28 induced by hypoxia-inducible factor-1a facilitated the migration of regulatory T cells to tumor sites in ovarian cancer, which contributed to the tumor tolerance and angiogenesis (15). Nevertheless, another study suggested that the reduced expression of CCL28 in colon tumors impaired the recruitment of IgA-secreting cells, which in turn, attenuated barrier function and promotes inflammation-driven tumor progression (35). Consistently, the results gained from the TCGA and Kaplan-Meier plotter cohorts validated the opposite prognostic values of CCL28 in luminal-like and triple-negative breast cancer, with no evidence showing an association between CCL28 and survival outcome in HER-2 enriched subtype. There existed a discrepancy about the prognostic effects of CCL28 among the three entire populations, which might partly be explained by the different sample sizes, baseline characteristics and cut-off values of these cohorts.
The divergent prognostic relevance of CCL28 might provide new insights into the clinical application of some emerging biomarkers for prediction of survival outcomes. Due to the significant difference of biological nature between subtypes in heterogeneous cancer, the same factor may play different or even opposite prognostic roles. Combining tumor subtypes with clinicopathologic features is of great significance to provide patients with comprehensive prognostic information.
To the best of our knowledge, the current study primarily performed a thorough investigation on the prognostic values of CCL28 in different subtypes of breast cancer with extended validation in large-sample datasets. However, this study has several limitations. First of all, given the retrospective nature, a powerful conclusion of the prognostic values of CCL28 could not be reached in this study. Further validation is warranted to be conducted on a prospective cohort with large sample size. Secondly, we measured the protein expression of CCL28 in the discovery cohort, but acquired the mRNA expression data of CCL28 from publicly available datasets. There must be an inconsistency between the protein and mRNA expression of CCL28. Moreover, the analyses based on the datasets consisting of gene expression data derived from various institutions and laboratories could inevitably introduce bias.
Overall, our study has shown for the first time that CCL28 served as a potential biomarker with complicated prognostic effect in luminal-like and triple-negative breast cancer. Future research on a prospective cohort with large sample size will further clarify the prognostic values of CCL28. As well, various involved pathways of CCL28 in different breast cancer subtypes deserve further exploration to better understand the role of CCL28 in breast cancer and the molecular signature of breast cancer by subtype.
Acknowledgements
Funding: This work was supported by the National Natural Science Foundation of China (No. 81722032, 81672600, 81370075), the Shanghai Shu-Guang Talent Project (No. 2016004) and the Ningbo Natural Science Foundation (No. 2017A610170).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: All procedures performed in this study were approved by the Ethical Committee of the Affiliated Cixi Hospital of Wenzhou Medical University (No. 050432-4-1212B), with all participants providing written informed consents.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Provenzano E, Ulaner GA, Chin SF. Molecular Classification of Breast Cancer. PET Clin 2018;13:325-38. [Crossref] [PubMed]
- Heneghan HM, Miller N, Lowery AJ, et al. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann Surg 2010;251:499-505. [Crossref] [PubMed]
- Yaal-Hahoshen N, Shina S, Leider-Trejo L, et al. The chemokine CCL5 as a potential prognostic factor predicting disease progression in stage II breast cancer patients. Clin Cancer Res 2006;12:4474-80. [Crossref] [PubMed]
- Dewan MZ, Ahmed S, Iwasaki Y, et al. Stromal cell-derived factor-1 and CXCR4 receptor interaction in tumor growth and metastasis of breast cancer. Biomed Pharmacother 2006;60:273-6. [Crossref] [PubMed]
- Teicher BA, Fricker SP. CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clin Cancer Res 2010;16:2927-31. [Crossref] [PubMed]
- Rollins BJ. Chemokines. Blood 1997;90:909-28. [PubMed]
- Balkwill F. Cancer and the chemokine network. Nat Rev Cancer 2004;4:540-50. [Crossref] [PubMed]
- Mantovani A, Savino B, Locati M, et al. The chemokine system in cancer biology and therapy. Cytokine Growth Factor Rev 2010;21:27-39. [Crossref] [PubMed]
- Li JY, Ou ZL, Yu SJ, et al. The chemokine receptor CCR4 promotes tumor growth and lung metastasis in breast cancer. Breast Cancer Res Treat 2012;131:837-48. [Crossref] [PubMed]
- Gu XL, Ou ZL, Lin FJ, et al. Expression of CXCL14 and its anticancer role in breast cancer. Breast Cancer Res Treat 2012;135:725-35. [Crossref] [PubMed]
- Wang W, Soto H, Oldham ER, et al. Identification of a novel chemokine (CCL28), which binds CCR10 (GPR2). J Biol Chem 2000;275:22313-23. [Crossref] [PubMed]
- Pan J, Kunkel EJ, Gosslar U, et al. A novel chemokine ligand for CCR10 and CCR3 expressed by epithelial cells in mucosal tissues. J Immunol 2000;165:2943-9. [Crossref] [PubMed]
- Hieshima K, Ohtani H, Shibano M, et al. CCL28 has dual roles in mucosal immunity as a chemokine with broad-spectrum antimicrobial activity. J Immunol 2003;170:1452-61. [Crossref] [PubMed]
- Facciabene A, Peng X, Hagemann IS, et al. Tumour hypoxia promotes tolerance and angiogenesis via CCL28 and T(reg) cells. Nature 2011;475:226-30. [Crossref] [PubMed]
- Dimberg J, Hugander A, Wagsater D. Protein expression of the chemokine, CCL28, in human colorectal cancer. Int J Oncol 2006;28:315-9. [PubMed]
- Mickanin CS, Bhatia U, Labow M. Identification of a novel beta-chemokine, MEC, down-regulated in primary breast tumors. Int J Oncol 2001;18:939-44. [PubMed]
- Yang XL, Liu KY, Lin FJ, et al. CCL28 promotes breast cancer growth and metastasis through MAPK-mediated cellular anti-apoptosis and pro-metastasis. Oncol Rep 2017;38:1393-401. [Crossref] [PubMed]
- Yu KD, Wang X, Yang C, et al. Host genotype and tumor phenotype of chemokine decoy receptors integrally affect breast cancer relapse. Oncotarget 2015;6:26519-27. [Crossref] [PubMed]
- Hao L, Zhang C, Qiu Y, et al. Recombination of CXCR4, VEGF, and MMP-9 predicting lymph node metastasis in human breast cancer. Cancer Lett 2007;253:34-42. [Crossref] [PubMed]
- Hammond ME, Hayes DF, Dowsett M, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol 2010;28:2784-95. [Crossref] [PubMed]
- Wolff AC, Hammond ME, Hicks DG, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol 2013;31:3997-4013. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61-70. [Crossref] [PubMed]
- Ciriello G, Gatza ML, Beck AH, et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell 2015;163:506-19. [Crossref] [PubMed]
- Gyorffy B, Surowiak P, Budczies J, et al. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One 2013;8:e82241. [Crossref] [PubMed]
- Gyorffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 2010;123:725-31. [Crossref] [PubMed]
- Goldhirsch A, Winer EP, Coates AS, et al. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol 2013;24:2206-23. [Crossref] [PubMed]
- TCGA Tissue Sample Requirements: High Quality Requirements Yield High Quality Data. Available online: https://cancergenome.nih.gov/cancersselected/biospeccriteria
- Liekens S, Schols D, Hatse S. CXCL12-CXCR4 axis in angiogenesis, metastasis and stem cell mobilization. Curr Pharm Des 2010;16:3903-20. [Crossref] [PubMed]
- Duda DG, Kozin SV, Kirkpatrick ND, et al. CXCL12 (SDF1alpha)-CXCR4/CXCR7 pathway inhibition: an emerging sensitizer for anticancer therapies? Clin Cancer Res 2011;17:2074-80. [Crossref] [PubMed]
- Tsang JYS, Ni YB, Chan SK, et al. CX3CL1 expression is associated with poor outcome in breast cancer patients. Breast Cancer Res Treat 2013;140:495-504. [Crossref] [PubMed]
- Nguyen AV, Wu YY, Liu Q, et al. STAT3 in epithelial cells regulates inflammation and tumor progression to malignant state in colon. Neoplasia 2013;15:998-1008. [Crossref] [PubMed]
- Muthuswamy RV, Sundstrom P, Borjesson L, et al. Impaired migration of IgA-secreting cells to colon adenocarcinomas. Cancer Immunol Immunother 2013;62:989-97. [Crossref] [PubMed]


