
Deep targeted sequencing analysis of hot spot mutations in non-small cell lung cancer patients from the Middle Eastern population
Introduction
Lung cancer is the leading cause of cancer deaths worldwide and in both men and women (1). Despite multiple novel therapeutic drugs, the overall 5-year survival remains around 15%. Non-small cell lung cancer (NSCLC) represents the majority (~80–85%) of diagnosed lung cancer cases. Lung adenocarcinoma (LUAD) and squamous cell carcinoma (LUSC) constitute the overwhelming majority of NSCLCs (2,3). While platinum based doublet therapy was considered the standard treatment for advanced stages for many years, accumulating evidence suggests that LUAD and LUSC, or even molecular subgroups of LUAD (e.g., KRAS-driven, EGFR-driven), represent different diseases that may benefit from disparate personalized therapies (4,5).
Multiple genetic and epigenetic alterations are involved in the tumorigenesis and the development of lung cancer. These cellular aberrations lead to constantly activated signaling pathways in cancer cells leading to uncontrolled cellular proliferation (5,6). The deciphering of the underlying pathogenesis and biological mechanisms and the increased availability of targeted therapeutics and biological markers has generated novel research and therapeutic avenues. In our current era of personalized medicine, the discovery of these targetable mutations in LUADs has paved the way for novel management strategies (7).
Over the last decade, testing for driver mutations in patients with LUAD has become routine practice especially with the presence of readily available therapeutic options for patients targeting some of these mutations, including epidermal growth factor receptor (EGFR), anaplastic lymphoma kinase (ALK), ROS1 and others. Data reported by the Lung Cancer Mutation Consortium (LCMC) have shown that patients with actionable mutations and actually receiving a targeted agent have a better survival when compared to patients not receiving the targeted agent or with no actionable mutation (8).
In current clinical practice, most centers rely on single-gene mutation testing to identify variants in genes like EGFR or ALK to guide clinicians to the most suitable therapy (3,4,9). Initial reports on the frequency of EGFR and ALK mutations in LUAD patients from the Middle East area have reported similar prevalence to Western populations. Shifting from single-gene mutation testing to simultaneous identification of the mutational landscape of a specific tumor is gaining ground and is being used more frequently in major academic medical centers (10-12). Next generation sequencing (NGS), particularly deep targeted sequencing, is currently a widely accepted methodology for identifying mutations in multiple cancer-related genes at the same time. NGS-based targeted sequencing has proved to be a satisfactory, clinically oriented assay to detect multi-genetic changes while using one platform (5,13). The original LCMC study tested 10 genes in samples from patients with metastatic LUADs simultaneously using multiplexed assays (8). Sixty-four percent (64%) of these patients had at least one mutation with KRAS, EGFR, and ALK being the most common ones (25%, 17%, and 8% respectively). Two similar prospective studies on NSCLC patients revealed that 54% and 51% of these patients had at least one mutation respectively (14,15).
While substantial differences in lung cancer genotypes are thought to exist amongst the different geographic regions, the molecular pathology of NSCLCs of Middle Eastern populations is poorly understood. We sought to begin to address this void by profiling canonical somatic cancer hot spot mutations in LUAD patients in the Middle East using deep targeted sequencing. In the present study, we characterized recurrent hot spot mutations in Middle Eastern LUAD (ME-LUAD). We also report that for some of these hot spots, their frequencies are dissimilar in ME-LUAD compared with LUADs from the West. Our finding lends support to the plausible supposition of a unique molecular pathology in ME-LUAD.
Methods
Tissue samples
Institutional Review Board approval was secured for this study and it conforms to the provisions of in accordance with the Helsinki Declaration (IRB number: IM.AT1.25). Patients with histologically confirmed LUADs were eligible for enrollment regardless of stage. Written informed consent was obtained from all patients. Patients were enrolled from eight sites in Lebanon, one in Iraq and one in Jordan. All patients signed informed consents. Demographic and clinic-pathological data were collected from all patients including age at diagnosis, gender, nationality, radiation exposure, medical history, tumor grade, stage, prior therapy, family history, and mutational status of EGFR assessed by Sanger sequencing. Specimens comprised formalin-fixed paraffin embedded (FFPE) core tissue biopsies either from the primary tumor or a metastatic site. Submitted slides were assessed for adequacy of tumor cells via histopathological assessment following hematoxylin and eosin staining.
DNA extraction
DNA extraction was performed on 85 FFPE tissue ribbons according to the Qiagen Manchester UK protocol at a College of American Pathologists-accredited laboratory at the American University of Beirut Medical Center (AUBMC) (16). The DNA quality and quantity were assessed using a Nanodrop ND-1000 spectrophotometer (Labtech, UK).
Sequencing and variant calling
Deep targeted sequencing using TruSeq Amplicon Cancer Panel (TSACP, 48 genes targeted with 212 amplicons, Illumina) and the MiSeq platform (Illumina) was conducted on 85 LUAD tumor tissue samples. Libraries consisting of 150 bp paired-end reads, were sequenced by the Avera Institute for Human Genetics at a median coverage depth of 3,056×. Raw reads quality check was performed using FastQC followed by adaptor removal and read trimming for low-quality calls (<15). Filtered sequence reads were aligned to human genome hg38 assembly using the Burrows-Wheeler Aligner, BWA program (17). Prior to somatic variant calling, we performed local realignment of the BWA-aligned reads using the Genome Analysis Toolkit (GATK) (14). For somatic variant calling, we used mutect2 from GATK on each sample independently with the local realigned reads from the previous step as input. Called variants were subsequently lifted over to hg19 for comparison with existing annotations (18).
Annotation was performed using the Variant Effect Predictor (VEP, v89) to classify variants into eight different classes (“Missense Mutation”, “Frame Shift Deletion”, “Frame Shift Insertion”, “In Frame Deletion”, “In Frame Insertion”, “Splice Site”, “Nonsense Mutation” and “Multi-Hit”) (16).Variants with an allele frequency >10% and a Sorting Intolerant From Tolerant (SIFT) score <0.05 (Deleterious) were retained. Oncoplots were generated from combining all samples and considering the top 20 mutated genes using maftools from Bioconductor (19,20). Clinical annotations including gender, age, smoking history, and EGFR mutation status from Sanger sequencing were integrated into the oncoplot.
Statistical analysis
The main outcome of this project was to examine the prevalence of hotspot mutations within a panel of 48 cancer-associated genes in LUADs patients evaluated in a tertiary care center in Lebanon and using a deep targeted sequencing assay. Descriptive statistics were used to present data on age at diagnosis, gender, nationality, prior radiation exposure, prior malignancy, smoking history, tumor grade, stage, prior therapy, EGFR mutation status and family history. The main dependent variables used were the mutation status of the top 10 mutated genes in our study categorized as either positive or negative. The independent variables used were age at diagnosis, gender, smoking history, tumor grade, and stage. Patients’ demographic, clinical and pathologic characteristics were compared to the presence or absence of a mutation using Pearson Chi-square tests or Fisher’s exact tests followed by odds ratio and 95% confidence intervals calculations. Analysis was performed using the statistical package IBM SPSS software version 24.0 (SPSS Inc., Chicago, IL, USA). P values <0.05 were considered statistically significant.
Multiplex PCR for EGFR mutational analysis
Tumor samples were analyzed for the presence of EGFR mutation by Reverse Transcription Polymerase Chain Reaction (RT-PCR), Amplification-refractory mutation system (ARMS) and Scorpion method on a RotorGene 3000 platform v2.0.2. EGFR PCR kits were used for specific mutations targeting exon 18 (G719 A, G719S, G719C), deletions in exon 19, exon 20 (T790M, S7681, and insertions), and exon 21 (L858R and L861Q).
Results
Deep targeted next-generation sequencing of 85 ME-LUADs
To gain insight into the molecular pathology of ME-LUAD, we used the TruSeq Amplicon-Cancer Panel (TSACP) designed to sequence mutational hotspots targeting 212 amplicons in 48 genes. The samples consisted of 93 FFPE tissues, of which eight failed sequencing quality check (QC) and 85 were retained for analysis (Figure 1A).
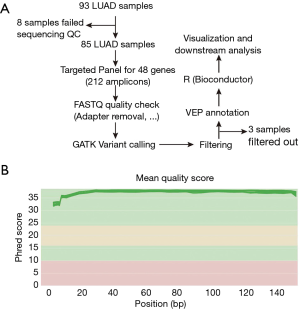
The remaining 85 samples consisted of 56 males and 29 females from five different neighboring countries in the Middle East (47 Lebanese, 15 Iraqis, 15 Jordanians, 6 Syrians, and 2 Palestinians) with a mean age at diagnosis of 63.7 years (Table 1). Sixty (70.6%) of our patients were either previous or current smokers with most of them presenting at stage IV (51.8%). The majority of the patients were wild type for EGFR following testing by Sanger sequencing (77/85; 90.6%) (Table 1).
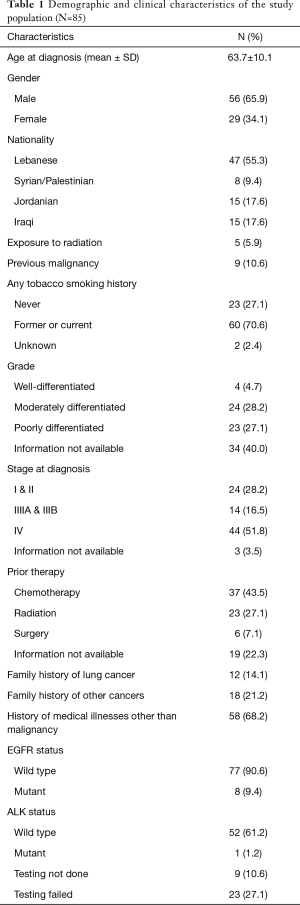
Full table
We performed deep paired-end sequencing on the 85 ME-LUADs with 150 read length on Illumina MiSeq platform leading to an average read count of 535,729 (Median of 501,461) per sample and an average depth of 3,056×. Quality check of sequenced reads reflected their high quality with an average quality score of 36.8 (Min =31.56; Figure 1B). Post-sequencing processing of samples and genomic analysis consisted of quality check, read filtering to remove adapter contamination, variant calling with GATK followed by variant filtering, annotation, and visualization (Figure 1; Methods).
Hotspot cancer-associated mutations in ME-LUAD
Variant calling identified a total of 2,455 variants, mostly missense mutations (1,855, 75.6%). We next filtered out germline mutations to focus on highly significant mutations, by removing variants with allele frequency (AF) <10% leaving those with a predicted deleterious effect on SNPs (SIFT score <0.05). This analysis yielded final set of 82 samples (Figure 2) with a total of 709 variants including 155 deletions (137 Frame shift and 18 In frame), 27 insertions (19 frame shift and 8 in frame) and 527 mutations composed of 519 missense mutations (73.2%), 1 nonsense mutation and 7 splice sites.

Driver mutations are responsible for a constantly activated signaling pathway leading to uncontrolled cell proliferation. In LUAD, several driver mutations have been identified with the 10 most common driver genes reported being EGFR, KRAS, ALK, ERBB2, BRAF, PIK3CA, MET, NRAS, AKT1, and TP53 (8). In our cohort, we found that all of our remaining 82 samples showed at least one mutation in the 10 most common driver genes in addition to FLT3, a class II receptor tyrosine kinase (Figure 2). Even though FLT3 was not in the TCGA cohort top 10 genes, it was the most commonly mutated gene in our LUAD cohort with 67% of samples affected. Notably, four of the above-mentioned genes (ALK, BRAF, NRAS, and AKT1) were not part of the top 20 most mutated genes (Figure 2). As for mutations in the BRAF gene, our study revealed that BRAF mutation frequency is very low, a finding common in the TCGA as well. Additionally, all samples with EGFR mutation screened independently by qPCR (Methods) showed concordant variants by deep targeted sequencing (Figure 2, red asterisk).
Mutational spectra and driver variants in ME-LUAD
Following assessment of hot spot mutations in the 85 ME-LUADs, we aimed at interrogating recurrent mutations in ME-LUAD. We pinpointed the most frequent mutations in our samples as well as cross-compared mutation frequencies between our ME population and Western cohorts. For this, we compared our samples mutation profile with mutations found for LUAD with a frequency >1% in TCGA (17) (Table 2).
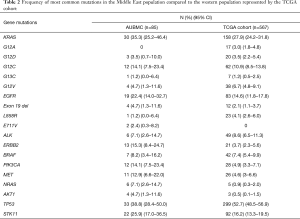
Full table
After FLT3, TP53, KRAS and STK11 were the first, second and third most commonly mutated genes, respectively, both in our cohort as well as in TCGA (Table 1). Of note, EGFR was mutated in 22.4% of our samples compared to 14.6% in TCGA.
The most prevalent mutation in LUAD, KRAS G12C, was found in 14.1% of our samples (95% CI: 7.5–23.4%) (Table 1) in accordance with its frequency in TCGA (10.9%). Similarly, for EGFR mutations, deletions in exon 19 were most prevalent (4.7%, 95% CI: 1.3–11.6%) and is similar to TCGA frequency (2.1%) (Table 2).
We then examined the correlation between the presence of a mutation in the top 10 mutated genes and demographic and clinical characteristics (age at diagnosis, gender, smoking status, and stage). Ninety Percent of KRAS-mutated cases were lifetime smokers (former or current) (P=0.006; OR 5.62, 95% CI: 1.50–20.99). We also noted that 91% of MET-mutated ME-LUADs were diagnosed in males (P=0.042; OR 1.45, 95% CI: 0.18–1.19). Notably, 77% of ME-LUADs with stage IV disease displayed TP53 mutations (P=0.001; OR 5.71, 95% CI: 2.05–15.92). Age was significantly associated with mutations in AKT1 and ERBB2. All AKT1-mutated samples belonged to patients who were diagnosed under the age of 60 (P=0.041; OR 0.9295% CI: 0.83–1.03]. Moreover, 84.9% of our tissue samples that were ERBB2-mutated represented adults who were over the age of 65 (P=0.024; OR 0.18, 95% CI: 0.03–0.90).
Discussion
The availability of targeted precision medicine has increased the interest of researchers in identifying the molecular alterations in patients with NSCLC (21,22). Detecting potentially actionable genetic mutations in NSCLC have been immensely aided by the development of NGS technologies. Our assay consisted of a panel of 48 genes in which some of the mutations in them have shown to affect both prognosis and response to therapy. The usage of NGS-based amplicon sequencing supports the implementation of this technique in the routine care of NSCLC instead of single-gene testing, and this is in line with several reports (5,9,23,24).
Identifying the molecularly-driven events in lung tumors is crucial in guiding therapy options (25,26). Genomic sequencing of a large set of DNA alterations in prior studies has shed light on the heterogeneity of lung cancers as well as its high mutational burden. In addition to the EGFR and ALK alterations, other mutations like KRAS, BRAF, ERBB2, and others are reported (27). Previous studies highlighted the presence of mutations in genes like PIK3CA, which can cause resistance to anti-EGFR or anti-ERBB2 therapy (22,28,29). The co-occurrence of tumor suppressor genes like TP53 with other oncogenes has been reported to impact the prognosis as well as the therapy (30-32). Moreover, MET amplification is also a known mechanism of resistance to anti-EGFR therapy (25,33). Other mutations involving AKT1 has the ability to influence patient’s response to inhibitors of EGFR/AKT pathway (34). This adds clinical value to the multi-gene testing made possible by using NGS-based platforms such as the one used in our study.
The frequencies of mutated genes presented in our study showed similarity to other reported outcomes across the spectrum of the available literature (23,35). At least 1 potentially actionable mutation was detected in all of our samples (100%) versus 60% in the LCMC cohort studied and 80% in Lindquist et al. (8,23). FLT3 was the most commonly mutated gene in our cohort followed by TP53 and KRAS (67%, 40%, and 37% respectively). FLT3 is known for its implication in acute myeloid leukemia and is reported to be amplified in only 0.4% in LUAD and mutated in 3.89% of 566 cases from TCGA and PanCancer studies (21,36). The high incidence of FLT3 mutations in our cohort may reflect in part false positive mutations possibly due to the lack of a reference genome for our Middle Eastern cohort.
TP53 was the most commonly mutated gene reported in a recent review combining data from genomic studies. Kadara et al. and The Cancer Genome Atlas (TCGA) also reported that the mutation rate of TP53 reached 43.5% and 45% respectively (21,22,25). Similarly, our data on KRAS mutation frequency is in line with literature report. The LCMC reported that KRAS was the most commonly mutated gene reaching 25% in their cohort of LUADs (8). Moreover, TCGA reported KRAS to reach 26.6% while it was 27.8% in Kadara cohort after TP53 (21,22). Another commonly mutated gene in LUAD is the EGFR gene which is worldwide reported around 15%. In our cohort of LUADs, the mutation rate reached 23% which is also higher than the reports highlighted in the above-mentioned large-cohort studies. In the literature, EGFR is reported to be more commonly mutated in lung tumors that belong to females and never-smokers (21-23,25). In our cohort of patients with LUADs, our never-smoking population reached a considerable 27.1% which explains the high EGFR frequency.
The results of the correlational analysis recapitulate the present reports on the mutational spectrum of LUADs. Patients with LUAD who ever-smoked commonly exhibit more somatic mutations as well as higher KRAS mutation frequency as exemplified in our results and other reports (21-23,25,35-37).
To the best of our knowledge, this is the first report on prospective collection of tumor tissue from 82 patients diagnosed with LUAD from 8 sites in Lebanon, 1 in Iraq and 1 in Jordan. In addition to that, the analysis of genetic alterations was performed in the context of treatment-naïve patients rather than post-therapy. This increases the sensitivity of our assays and the validity of the results especially for EGFR mutations. Even though there are current commercial assays for multi-gene testing present, the technique used in our study does not only focus on hotspot alterations in specifically-identified set of genes but also provides quantitative variant measurement as well as simultaneous detection of concurring mutations and insertions/deletions. In addition to that, the amount of DNA material needed for NGS is less which enables researchers to widen their scope of testing. Utilizing NGS-based platforms increases the options of actionable mutations and targeted therapies available for each patient while keeping the cost, time and effort consumed comparable to single-gene testing.
Paired healthy tissue from the same patient with LUAD was not provided for comparative analysis and this is one of the limitations of our project. The presence of both datasets would have decreased our false positive rate and increased our specificity. On another note, although the sample size sounds large, it has a relatively diverse genetic, geographical, cultural and lifestyle backgrounds increasing thus variability and affecting our analysis by adding several confounding factors influencing our samples predisposition to lung cancer. More trials are encouraged to use NGS-based platforms to assay genetic mutations and link them to their interventional therapies. NGS-based platforms are only newly being used to assay molecular and genetic alterations. Thus, it may seem too early to judge on its clinical impact on choice and response of therapy using follow-up studies. However, in the future, it will be important to confirm that the correct treatment options for patients have been made.
Accurate diagnostic testing for lung cancer is crucial because of its high impact on prognosis and choice of therapy. In summary, despite the fact that many of the genes tested did not reach high mutation frequencies, at least one mutation was detected in all of the samples. Acknowledging that the worldwide population’s incidence of lung cancer is increasing, our results support the routine testing of these genes in screening programs or clinical trials to aide clinicians in providing the most suitable individualized treatment for each patient. The spectrum of commercially-available targeted therapies remains limited thus deeming large comprehensive genetic evaluation excessive especially because of the high cost it incurs on the patient.
Acknowledgments
This work was supported by an intramural funding source from Medical Practice Plan (MPP) from the American University of Beirut.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by Institutional Review Board (No. IM.AT1.25) and written informed consent was obtained from all patients.
References
- Cancer. World Health Organization. Available online: http://www.who.int/mediacentre/factsheets/fs297/en/. Accessed December 1, 2018.
- Sequist LV, Heist RS, Shaw AT, et al. Implementing multiplexed genotyping of non-small-cell lung cancers into routine clinical practice. Ann Oncol 2011;22:2616-24. [Crossref] [PubMed]
- Schiller JH, Harrington D, Belani CP, et al. Comparison of Four Chemotherapy Regimens for Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2002;346:92-8. [Crossref] [PubMed]
- Oxnard GR, Binder A, Jänne PA. New Targetable Oncogenes in Non-Small-Cell Lung Cancer. J Clin Oncol 2013;31:1097-104. [Crossref] [PubMed]
- Scarpa A, Sikora K, Fassan M, et al. Molecular Typing of Lung Adenocarcinoma on Cytological Samples Using a Multigene Next Generation Sequencing Panel. PLoS One 2013;8. [Crossref] [PubMed]
- Sato M, Shames DS, Gazdar AF, et al. A Translational View of the Molecular Pathogenesis of Lung Cancer. J Thorac Oncol 2007;2:327-43. [Crossref] [PubMed]
- Miller YE. Pathogenesis of Lung Cancer. Am J Respir Cell Mol Biol 2005;33:216-23. [Crossref] [PubMed]
- Kris MG, Johnson BE, Berry LD, et al. Using Multiplexed Assays of Oncogenic Drivers in Lung Cancers to Select Targeted Drugs. JAMA 2014;311:1998. [Crossref] [PubMed]
- Hagemann IS, Devarakonda S, Lockwood CM, et al. Clinical next-generation sequencing in patients with non-small cell lung cancer. Cancer 2015;121:631-9. [Crossref] [PubMed]
- Herbst RS, Heymach JV, Lippman SM. Lung Cancer. N Engl J Med 2008;359:1367-80. [Crossref] [PubMed]
- Cardarella S, Ortiz TM, Joshi VA, et al. The introduction of systematic genomic testing for patients with non-small cell lung cancer. J Thorac Oncol 2012;7:1767-74. [Crossref] [PubMed]
- Tfayli A, Khalil M, Mina A, et al. Screening for the Prevalence of EGFR and ALK Mutations in Lung Adenocarcinoma Patients in The Levant Area: A Prospective Analysis. Asian Pac J Cancer Prev 2017;18:107-14. [PubMed]
- Gailey MP, Stence AA, Jensen CS, et al. Multiplatform comparison of molecular oncology tests performed on cytology specimens and formalin-fixed, paraffin-embedded tissue. Cancer Cytopathol 2015;123:30-9. [Crossref] [PubMed]
- Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst 2005;97:339-46. [Crossref] [PubMed]
- Shamseddine A, Sibai AM, Gehchan N, et al. Cancer incidence in postwar Lebanon: findings from the first national populationbased registry. Ann Epidemiol 2004;14:663-8. [Crossref] [PubMed]
- Therascreen® EGFR RGQ PCR Kit Handbook. Version 2. Qiagen Manchester Ltd, Skelton House, Lloyd Street North, Manchester, M15 6SH, UK. REF 874111. August 2016.
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754-60. [Crossref] [PubMed]
- Cibulskis K, Lawrence MS, Carter SL. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213-9. [Crossref] [PubMed]
- McLaren W, Gil L, Hunt SE, et al. The Ensembl Variant Effect Predictor. Genome Biology 2016;6;17:122.
- Mayakonda A, Koeffler HP. Maftools: Efficient analysis, visualization and summarization of MAF files from large-scale cohort based cancer studies. BioRxiv. 2016.
- The Cancer Genome Atlas Network A. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref]
- Kadara H, Choi M, Zhang J, et al. Whole-exome sequencing and immune profiling of early-stage lung adenocarcinoma with fully annotated clinical follow-up. Ann Oncol 2017;28:75-82. [PubMed]
- Lindquist KE, Karlsson A, Levéen P, et al. Clinical framework for next generation sequencing based analysis of treatment predictive mutations and multiplexed gene fusion detection in non-small cell lung cancer. Oncotarget 2017;8. [Crossref] [PubMed]
- Boland GM, Piha-Paul SA, Subbiah V, et al. Clinical next generation sequencing to identify actionable aberrations in a phase I program. Oncotarget 2015;6. [Crossref] [PubMed]
- Devarakonda S, Morgensztern D, Govindan R. Genomic alterations in lung adenocarcinoma. Lancet Oncol 2015;16:e342-51. [Crossref] [PubMed]
- Zhang J, Fujimoto J, Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science 2014;346:256-9. [Crossref] [PubMed]
- Pao W, Hutchinson KE. Chipping away at the lung cancer genome. Nat Med 2012;18:349-51. [Crossref] [PubMed]
- Hanker AB, Pfefferle AD, Balko JM, et al. Mutant PIK3CA accelerates HER2-driven transgenic mammary tumors and induces resistance to combinations of anti-HER2 therapies. Proc Natl Acad Sci USA 2013;110:14372-7. [Crossref] [PubMed]
- Majewski IJ, Nuciforo P, Mittempergher L, et al. PIK3CA mutations are associated with decreased benefit to neoadjuvant human epidermal growth factor receptor 2-targeted therapies in breast cancer. J Clin Oncol 2015;33:1334-9. [Crossref] [PubMed]
- Pécuchet N, Laurent-Puig P, Mansuet-Lupo A, et al. Different prognostic impact of STK11mutations in non-squamous non-small-cell lung cancer. Oncotarget 2017;8. [Crossref] [PubMed]
- Huang S, Benavente S, Armstrong EA, Li C, Wheeler DL, Harari PM. p53 Modulates Acquired Resistance to EGFR Inhibitors and Radiation. Cancer Res 2011;71:7071-9. [Crossref] [PubMed]
- Ma X, Teuff GL, Lacas B, et al. Prognostic and Predictive Effect of TP53 Mutations in Patients with Non-Small Cell Lung Cancer from Adjuvant Cisplatin-Based Therapy Randomized Trials: A LACE-Bio Pooled Analysis. J Thorac Oncol 2016;11:850-61. [Crossref] [PubMed]
- Camidge DR, Pao W, Sequist LV. Acquired resistance to TKIs in solid tumours: learning from lung cancer. Nat Rev Clin Oncol 2014;11:473-81. [Crossref] [PubMed]
- Carpten JD, Faber AL, Horn C, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature 2007;448:439-44. [Crossref] [PubMed]
- Barlesi F, Mazieres J, Merlio JP, et al. Routine molecular profiling of patients with advanced non-small-cell lung cancer: results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT). Lancet 2016;387:1415-26. [Crossref] [PubMed]
- Gahr S, Stoehr R, Geissinger E, et al. EGFR mutational status in a large series of Caucasian European NSCLC patients: data from daily practice. Br J Cancer 2013;109:1821-8. [Crossref] [PubMed]
- Hammerman PS, Lawrence MS, Voet D, et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519-25. [Crossref] [PubMed]


