
Application of radiomics signature captured from pretreatment thoracic CT to predict brain metastases in stage III/IV ALK-positive non-small cell lung cancer patients
Introduction
Lung cancer is one of the most common malignant tumors and it remains the leading cause of death worldwide (1,2). Among all pathological types of lung cancer, non-small cell lung cancer (NSCLC) accounts for 85%. Approximately 30–43% of NSCLC patients have brain metastases (BM) when the disease progresses (3,4). Once BM emerges, the natural median progression-free survival (PFS) is only 1–2 months, and the 1-year survival rate is as low as 10–20% (5). More than 60% lung adenocarcinoma has driver gene mutations, among which epidermal growth factor receptor (EGFR) mutation is one of the most important, with incidence of 30% in Asian (6). Approximately 3–5% NSCLC have rearrangements in anaplastic lymphoma kinase (ALK-positive), and echinoderm microtubule-associated protein like 4-anaplastic lymphoma kinase (EML4-ALK) fusion is the most frequently observed (7,8). Despite the obvious difference in their occurrence probability, the incidence of brain metastasis (BM) in ALK-positive (tyrosine-kinase inhibitor-naive) patients (20–35%) is comparable to that observed in EGFR-mutated NSCLC patients (9,10). For ALK-positive lung cancer patients treated with TKIs, BM is the most common pattern of failure (60%) (11). Therefore, BM status might be significant in evaluating patients’ prognosis and curative effect, and it is necessary to explore effective approaches to predict it.
The suffix “-omics” is now widely applied in research of cancers to demonstrate the idea of extracting high-throughput data from tumors. Genomics is defined as a discipline concerned with investigation of structure and function of genomes (12). Driver gene status of lung cancer, such as EML4-ALK fusion mentioned above, demonstrates tumor heterogeneity from microscopic molecular levels. In 2003, Baumann et al. (13) proposed the GENEPI project, which gave birth to the concept of “radiogenomics”. The original definition of radiogenomics was limited to the prediction of radiosensitivity based on tumor gene expression (14). Then researchers began to analyze the relationship between gene expression and “radiomics” features, thus enlarging the extent and depth of radiogenomics (15,16).
Radiomics refers to high-throughput extraction of quantitative image features that provide a comprehensive description of tumor phenotypes and heterogeneity (17-20).
Given the unprecedented presence of radiomics by Lambin et al. in 2012, the exploration and extended application of radiomics has developed very fast recently, especially in the field of lung cancer (21-23). Biomarkers based on radiomics features can be related to different clinical outcomes and potential genomic phenotypes, which is meaningful for risk assessment and prognostic evaluation (24). Yoon et al. discovered a radiomics approach that could decode different tumor phenotypes of ALK, C-ros oncogene 1 receptor tyrosine kinase (ROS1), and RET fusions in lung adenocarcinoma (25), which implies that radiomics could be applied in ALK-positive NSCLC patients.
Radiomics has been reported to be valuable in predicting lymph node metastases in different kinds of tumors (26-29), which indicates that radiomics has the potential to be developed into a marker of tumor metastasis. As far as we know, however, there have not been abundant reports regarding applying radiomics methods in predicting BM for ALK-positive NSCLC patients. Under this premise, we seek to develop a radiomics approach to predict BM for ALK-positive NSCLC patients in this study, which allows the possibility for radiomics to guide individualized risk assessment for such group of patients.
Methods
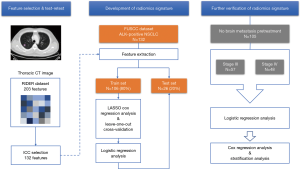
Our study was conducted according to the flow chart in Figure 1.
Data description
NSCLC patients with pathologically confirmed ALK rearrangement from June 2014 to September 2017 in Fudan University Shanghai Cancer Center (FUSCC) were enrolled retrospectively in this study. There were 366 patients in total before exclusion. This retrospective study strictly obeyed the principles of the Declaration of Helsinki. This retrospective study was approved by the Fudan University Shanghai Cancer Center Institutional Review Board (No. 1904199-14-1905) and all methods were performed in accordance with the guidelines and regulations of this ethics board. All participants signed their informed consent after being fully informed of the purpose and content of this study. Patients included in this study met the following qualifications:
- All patients were staged according to the 7th Edition of AJCC Cancer Staging Manual [2010], and those who were staged by medical images as overall stage III/IV NSCLC were enrolled.
- ALK rearrangement status was detected by immunohistochemistry (IHC) and fluorescent in situ (FISH) on tumor tissues in FUSCC, and those with ALK-positive NSCLC were qualified for this study.
- All patients’ thoracic CT images and RT structure at baseline examination could be collected from the Pinnacle system (8.0 m Philips Healthcare, Andover, MA, USA) and MIM system (version 6.6, MIM Software Inc. Cleveland, Ohio, USA).
- All patients’ medical records, including treatment information and surveillance examinations, could be collected from the Electronic Medical Records System (EMRS) at FUSCC.
Clinical observation
The primary endpoint of this study was BM, which was defined as progression of disease to brain as assessed by baseline or surveillance brain MRI/CT scans both in and out of FUSCC. Time to BM was defined as time from the diagnosis of NSCLC to the date of BM or censoring (date of last scan or follow-up).
Clinical variables and treatment information
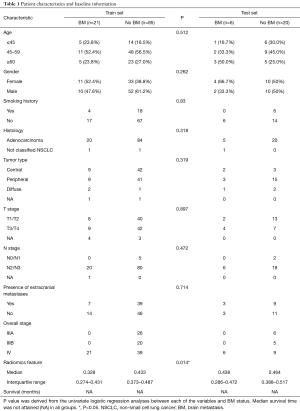
The baseline characteristics (e.g., histology, tumor type, overall stage, etc.) were recorded, and conventional clinical prognostic factors (CPFs) used for this study included age (1= ≤45, 2= 45–59, 3= ≥60), gender (0= male, 1= female), smoking history (0= no, 1= yes), T stage (1= stage 1–2, 2= stage 3–4), N stage (1= stage 0–1, 2= stage 2–3) and presence of extracranial metastasis (0= no, 1= yes). Treatment elements during follow-up were recorded (e.g., whether receive surgery, radiotherapy, chemotherapy and target therapy or not).
CT acquisition and segmentation
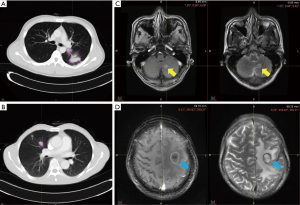
Pretreatment chest CT images were acquired with a Siemens Somatom Sensation MSCT scanner (Siemens Healthineers, Erlangen, Germany) according to the standard scanning protocol applied in FUSCC. In this study, we used contrast enhanced thoracic CT images. CT scan parameters were as follows: tube voltage, 120 kV; automatic tube current modulation, 200 mAs; matrix, 512×512. The scan ranged from the tip of the lung to the base of the lung, including both axillae. The original data were reconstructed by 1 mm thin layer and multiple plane reconstruction. The CT images had not been normalized. The gross tumor volume (GTV) was delineated by one radiation oncologist and reviewed by another senior radiation oncologist on the MIM system and both radiation oncologists had more than ten years of clinical experience in thoracic cancer. The boundaries between chest and other tissues were primarily identified at the abdomen window, leaving the lung window for contouring (Figure 2A,B).
Feature extraction and test-retest operation
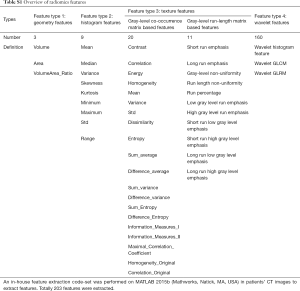
In this study, we performed an in-house feature extraction code-set based on MATLAB 2015b (Mathworks, Natick, MA, USA) in patients’ CT images (30). In total, 203 features from CT scans were extracted, which could be divided into seven subgroups (Table S1 and supplementary material):
- Wavelet gray level co-occurrence matrix-based features;
- Wavelet gray level run-length matrix-based features;
- Wavelet histogram-based features;
- Gray level co-occurrence matrix-based features;
- Gray level run-length matrix-based features;
- Geometry features;
- Histogram features.
Full table
In feature extraction, we used DICOM data. Gray level rescaling was defined by DICOM, 100 gray levels. As for texture features, we used 2D texture matrix, four directions in 2D offset, and features were generated by averaging over all offsets. For wavelet features, we used a multilevel 2-D stationary wavelet decomposition, which was the ‘swt2’ function in MATLAB. The wavelet filter belongs to ‘coiflet’ wavelet family. Here we used ‘coif1’.
Regarding RIDER NSCLC date, a detailed CT protocol and segmentation were available from Aerts et al. (31) and Zhao et al. (32). In our study, we employed a RIDER test-retest dataset to identify the most stable features between two interval CT scans (33). The primary tumor was segmented both in test scans and retest scans by applying the Grow Cut algorithm running on the 3D Slicer (version 4.7.0, USA). The intraclass correlation coefficient (ICC) was utilized to measure fixity. We performed a two-way fixed effect, absolute agreement, and single measurement model for ICC calculations (34). ICC values range from 0 to 1, and values closer to 1 indicate better stability.
Feature selection
All patients enrolled in this study were divided into a training set and test set before treatment (4:1). The least absolute shrinkage and selection operator (LASSO) Cox regression analysis with a leave-one-out cross-validation was performed on the training set to identify the most suitable and adequate radiomics model by reducing parameter redundancy.
Development and validation of radiomics signature in predicting pre-treatment BM for ALK-positive NSCLC patients
Firstly, we tried to develop a predictive signature for predicting pretreatment BM for ALK-positive NSCLC patients (Figure 2C). To achieve this goal, the logistic regression model was trained for the radiomics feature(s) and other traditional CPFs on a training set, and the test set was used as an independent validation cohort to verify the performance of the signature. The receiver operating characteristic (ROC) curve was constructed to assess the predictive power of the signature, and the area under curve (AUC) was calculated.
Radiomics signature-based predictive analysis of BM during follow-up observation
For those without BM at baseline assessment, we then assessed the predictive ability of the signature for the emergence of BM during/after treatment (Figure 2D). Considering different treatment strategy and highly variable prognosis, those patients were divided into stage III and stage IV groups for further analyses. A ROC analysis was performed for each group.
Assessment of prognostic value of general signature for BM
After proving the extended predictive value of the radiomics signature during surveillance, we then further explored the possibility of using this approach to develop a stratified model for BM during surveillance. For these locally advanced and advanced NSCLC patients, treatment strategy was individualized because of patients’ general condition and preference. So, treatment features were also taken into consideration (e.g., whether receive surgery, radiotherapy, chemotherapy and target therapy or not). We conducted Cox regression analysis to evaluate the prognostic performance of the radiomics signature and potential treatment features for BM. The general index (Gen_index) was calculated for each patient via a linear combination of features multiplied by their coefficients. Patients were classified into high risk or low risk groups according to Gen_index, and the appropriate threshold was determined by using the median value of Gen_index. Log-rank test was used to describe the significance of difference between two groups.
Data analysis
A test-retest analysis was performed to identify stable radiomics features. LASSO Cox regression and a leave-one-out cross-validation were conducted to reduce redundant. Variables’ relationship with pretreatment BM status was measured by univariable logistic regression analyses. Performance of logistic regression model was measured by AUC of ROC curve. Statistical comparison between group III and IV was performed using Chi Square test. Cox regression analyses were performed to find prognostic clinical and radiomics feature(s) for BM during follow-up and log-rank test was performed to describe the difference between two risk groups. Statistical analyses were performed using R software (version 3.3.2; http://www.Rproject.org), the reported statistical significance levels were all two-sided, with statistical significance set at 0.05.
Results
Clinical data
In total, 132 patients were enrolled retrospectively, including 27 patients with BM at baseline examination. The median follow-up time was 11.8 months (range, 0.1–65.2 months). All patients were randomly divided into two groups: training set (80%) and test set (20%). The median survival time had not been attained (NA) yet in both sets. The training set included a total of 106 patients, and 21 of them had BM before treatment. Regarding the 26 patients in the test set, totally six of them had pretreatment BM. Detailed baseline information is provided in Table 1.
Full table
Test-retest reliability and construction of radiomics signature
Among all 203 features in the RIDER dataset, we identified 132 stable features (65%) with an ICC greater than 0.9, which was considered to indicate high reliability. We then included the 132 features into LASSO regression model for developing an adequate radiomics signature to predict pretreatment BM. Only one feature was qualified for the radiomics signature: W_GLCM_LH_Correlation (P value =0.014).
Performance of radiomics signature in predicting pretreatment BM in ALK-positive NSCLC patients
No clinical variable was found significantly predictive of pretreatment BM in the training set except the radiomics feature (W_GLCM_LH_Correlation). The detailed information of univariate logistic regression is presented in Table 1. The predictive model was defined as:
logit (P) = 0.819-5.696× W_GLCM_LH_Correlation
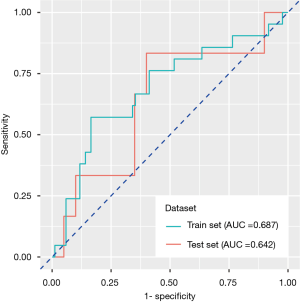
In the training set, the predictive power of the model was measured by the area under the ROC curve, with an AUC of 0.687 (95% CI: 0.551–0.824), specificity of 83.5% and sensitivity of 57.1%, and the model also exhibited modest performance in the test set, with an AUC of 0.642 (95% CI: 0.501–0.783), specificity of 60.0% and sensitivity of 83.3% (Figure 3).
Application of radiomics signature in predicting BM in follow-up observation
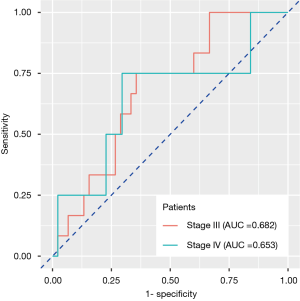
Those 105 patients without BM at baseline examination were divided into groups of stage III (n=57) and stage IV (n=48). Stage IV patients refer to those harboring extracranial metastasis at baseline examination. We continued monitoring their BM status during follow-up. Twelve stage III and four stage IV patients had BM, respectively, during or after treatment. The radiomics signature developed previously also exhibited reposeful performance to predict BM during follow-up in two groups separately (stage III: AUC =0.682, 95% CI: 0.537–0.826, specificity =64.4%, sensitivity =75.0%; stage IV: AUC =0.653, 95% CI: 0.503–0.804, specificity =70.4%, sensitivity =75.0%), implying that the feature correlated with pretreatment BM also had relatively stable predictive value in the follow-up observation (Figure 4).
Prognostic value and risk stratification ability of general signature
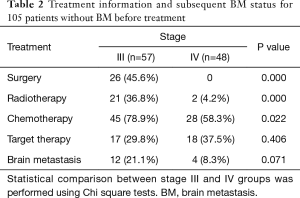
The treatment information for the 105 patients until the emergence of BM (event) or until the date that the patient was last known to be free from BM is presented in Table 2.
Full table
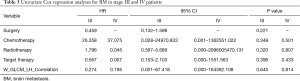
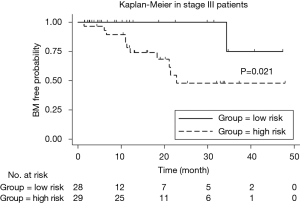
We implemented univariate Cox regression analyses to assess the prognostic value of radiomics and treatment features associated with BM during surveillance for two groups. The results are presented in Table 3. Neither radiomics feature nor treatment features were significantly prognostic for BM in either stage III or IV patients (P value >0.05). However, in the stage III group, the multivariate Cox regression model which incorporated variables of whether receive chemotherapy/radiotherapy or not and the radiomics feature, was able to identify high risk or low risk patients for BM by stratification analysis (log-rank P value =0.021). Mean BM free time for high risk group was 30.6 (95% CI: 23.2–38.1) months. Mean BM free time for low risk group was 44.0 (95% CI: 38.6–49.5) months (Figure 5).
Full table
Discussion
For most lung cancer patients, disease is locally advanced (stage III) or advanced (stage IV) at the time of diagnosis.For these patients, some stage III patients are eligible for surgery, whereas most of them need diversified treatment strategies based on radiotherapy and chemotherapy. Although these traditional treatment strategies are effective to some extent, the overall therapeutic outcomes remain unsatisfying. One of the leading causes might be neglect of tumor heterogeneity. Estimating the risk of invasion and recurrence according to different genotypes and phenotypes of lung cancer will be the key to comprehensively understanding the heterogeneity of tumors and implementing individualized treatment. Great efforts have been contributed to the investigation of this issue in recent years (6,35-38).
In microscopic terms, tumor heterogeneity is traditionally measured and differentiated based on tissue cytology and gene level. Approximately 5% of NSCLC patients harbor ALK rearrangements, brain metastases and disease progression in the brain are very common in ALK-positive patients (39). Despite the increasing number of investigations seeking to establish specific predictive and prognostic models for ALK-positive NSCLC patients (35), an effective and acknowledged method for predicting BM in such a group of patients has not been reported yet. Thus, our research is enlightening and exhibits potential for future studies.
Radiomics describes tumor heterogeneity from a macroscopic perspective. Numerous studies on its predictive value for tumor metastases and progression have been performed in recent years, and most of them focused on lymphatic metastasis of various types of tumors (26,29,40-42), including lung cancer (27,28). Nevertheless, research using a radiomics approach based on thoracic CT images in the prediction of BM among ALK-positive NSCLC patients has been rarely reported to date, so our study is original and novel. A recent study by Chaddad et al. (43) proved that quantitative thoracic CT imaging features may serve as indicators of survival for patients with large-cell-carcinoma (LCC), primary-tumor-sizes (T2) and no lymph-node-metastasis (N0). The result of our study was consistent with Chaddad et al.’s in terms of proving radiomics feature’s capability in risk prediction and prognostic evaluation for NSCLC patients. However, our study focused on a minority of NSCLC patients with high incidence of BM and we aimed at finding possible radiomics marker for predicting BM status for such patients, which has positive meaning for clinical treatment decision-making and prognosis evaluation. In this sense, our study has made positive attempt in extending radiomics application space in predicting distant metastasis.
In this study, we analyzed 132 radiomics features out of 203 features extracted from pretreatment thoracic CT images of stage III/IV NSCLC patients harboring EML4-ALK fusion, and one radiomics feature (W_GLCM_LH_Correlation) was found predictive of BM before treatment. Gray level co-occurrence matrix (GLCM) is one type of texture feature that describes the values, distances, and angles of gray combinations of image (44), and W_GLCM_LH_Correlation is a wavelet transformed texture feature. Among all radiomics features, texture features are known to be most closely related with tumor heterogeneity and prognosis, while wavelet feature is the result of filter transformation of intensity and texture features (24). Some studies have successfully developed texture and wavelet features to distinguish NSCLC from benign lung lesions (14). Radiomics approach based on texture and wavelet features can serve as predictive markers of EGFR, Kirsten rat sarcoma viral oncogene (KRAS), ALK and ROS1 gene mutations in lung cancer (25,45). Previous studies have also confirmed that radiomics features could predict local recurrence or distant metastasis of lung cancer. Coraller et al. (46) constructed a radiomics model with 635 features, and 35 of them can predict distant metastasis, mainly including two types: wavelet HHL-Skewness and texture feature GLCM-Cluster shade. Ferreira Junior et al. (47) found that radiomics characterization approach presented great potential in lymph node metastasis, distant metastasis, and histopathology pattern recognition. Our study further confirmed that wavelet transformed texture feature can be related to early BM in lung cancer patients with specific driver gene mutation (ALK-positive). The conclusion of this study is also consistent with the previous study. Regarding acknowledged CPFs, such as smoking history (46), none were significantly correlated with BM in this study, indicating that ALK-positive NSCLC might be a unique subtype of NSCLC and it is pressing to identify unique predictive methods for such patients.
Retrospective analysis of ALK-positive NSCLC patients with BM enrolled in PROFILE 1005 and PROFILE 1007 (11) demonstrates that crizotinib contributes to a high disease control rate for BM. Nevertheless, brain remains the most common site of progression in patients with or without baseline BM. Thus, it is essential to develop predictable and prognostic indicators for BM not only before treatment but also during surveillance. We verified the further predictive value of the radiomics signature for BM emergence during follow-up for those without BM at baseline examination, and we eventually observed its promising performance in separate groups of stage III and stage IV patients, indicating that the radiomics feature has the potential to represent a stable biomarker correlated with BM in long-term surveillance. No significant discovery resulted from individually introducing clinical treatments covariates into the prognostic evaluation of BM in either group. However, the integration of radiomics feature and treatments elements (chemotherapy and radiotherapy), was helpful to risk assessment for BM in stage III patients. It is well known that concurrent and sequential chemoradiotherapy are standard treatment strategies for stage III NSCLC patients, and our study confirms that chemoradiotherapy for such patients may have some reference for the emergence of BM during follow-up observation.
There are some limitations in this study. Considering the general low incidence of ALK rearrangement among NSCLC patients and that many patients had already received various therapies before they came to FUSCC, the sample size of this study is limited. An expanded sample size and external multicenter validation are necessary for further investigations in order to verify the results of this study. In addition, it is necessary to include NSCLC patients with other driver gene mutations (e.g., EGFR, ROS1 and KRAS) to explore the extensibility and universal applicability of this radiomics signature. Finally, our study was implemented on chest CT images, and the predictive value of the radiomics signature could be further explored in other clinical images, such as magnetic resonance imaging (MRI) of the brain or positron emission tomography (PET), which are both essential examinations at baseline evaluation for NSCLC patients.
Conclusions
In conclusion, we discovered that a radiomics wavelet texture feature W_GLCM_LH_Correlation, which was derived from pretreatment thoracic CT, presented potential in predicting BM in stage III/IV ALK-positive NSCLC patients. This preliminary finding allows possibility in exploring risk prediction models for early identification of BM for such patients.
Supplementary
Radiomics features extraction
An in-house feature extraction code-set was performed on MATLAB 2015b (Mathworks, Natick, MA, USA) in patients’ CT images to extract features. Totally 203 features were extracted (Table S1). Following equations define some of these features.
GLCM features
Notation:
p(i,j): (i,j)th entry in a normalized gray-tone spatial-dependence matrix, =P(i,j)/R
Ng: Number of distinct gray levels in the quantized image.
px(i): ith entry in the marginal-probability matrix obtained by summing the rows of p(i,j),
| [1] |
| [2] |
| [3] |
Contrast:
| [4] |
Correlation:
| [5] |
Where μx, μy, σx, σy are the means and standard deviations of px(i) and py(j).
Variance:
| [6] |
Entropy:
| [7] |
Sum average:
| [8] |
Sum variance:
| [9] |
Sum entropy:
| [10] |
Difference variance:
| [11] |
Difference entropy:
| [12] |
Information measures of correlation:
| [13] |
| [14] |
| [15] |
Where HX and HY are entropies of px and py, and
| [16] |
| [17] |
Maximal correlation coefficient:
| [18] |
Where
| [19] |
GLRLM feature
Notation:
| [20] |
| [21] |
Short run emphasis (SRE):
| [22] |
Long run emphasis (LRE):
| [23] |
Gray-level nonuniformity (GLN):
| [24] |
Run length nonuniformity (RLN):
| [25] |
Run percentage (RP):
| [26] |
In the above, nr is the total number of runs and np is the number of pixels in the image.
Low gray-level run emphasis (LGRE):
| [27] |
High gray-level run emphasis (HGRE):
| [28] |
Short run low gray-level emphasis (SRLGE):
| [29] |
Short run high gray-level emphasis (SRHGE):
| [30] |
Long run low gray-level emphasis (LRLGE):
| [31] |
Long run high gray-level emphasis (LRHGE):
| [32] |
Acknowledgments
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This retrospective study strictly obeyed the principles of the Declaration of Helsinki. This retrospective study was approved by the Fudan University Shanghai Cancer Center Institutional Review Board (No.1904199-14-1905) and all methods were performed in accordance with the guidelines and regulations of this ethics board. All participants signed their informed consent after being fully informed of the purpose and content of this study.
References
- Chen W, Zheng R, Zhang S, et al. Cancer incidence and mortality in China in 2013: an analysis based on urbanization level. Chin J Cancer Res 2017;29:1-10. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Nayak L, Lee EQ, Wen PY. Epidemiology of brain metastases. Curr Oncol Rep 2012;14:48-54. [Crossref] [PubMed]
- Olmez I, Donahue BR, Butler JS, et al. Clinical outcomes in extracranial tumor sites and unusual toxicities with concurrent whole brain radiation (WBRT) and Erlotinib treatment in patients with non-small cell lung cancer (NSCLC) with brain metastasis. Lung Cancer 2010;70:174-9. [Crossref] [PubMed]
- Tabouret E, Chinot O, Metellus P, et al. Recent trends in epidemiology of brain metastases: an overview. Anticancer Res 2012;32:4655-62. [PubMed]
- Fukuoka M, Wu YL, Thongprasert S, et al. Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS). J Clin Oncol 2011;29:2866-74. [Crossref] [PubMed]
- Gainor JF, Varghese AM, Ou SH, et al. ALK rearrangements are mutually exclusive with mutations in EGFR or KRAS: an analysis of 1,683 patients with non-small cell lung cancer. Clin Cancer Res 2013;19:4273-81. [Crossref] [PubMed]
- Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. [Crossref] [PubMed]
- Doebele RC, Lu X, Sumey C, et al. Oncogene status predicts patterns of metastatic spread in treatment-naive nonsmall cell lung cancer. Cancer 2012;118:4502-11. [Crossref] [PubMed]
- Rangachari D, Yamaguchi N, VanderLaan PA, et al. Brain metastases in patients with EGFR-mutated or ALK-rearranged non-small-cell lung cancers. Lung Cancer 2015;88:108-11. [Crossref] [PubMed]
- Costa DB, Shaw AT, Ou SH, et al. Clinical experience with crizotinib in patients with advanced ALK-rearranged non-small-cell lung cancer and brain metastases. J Clin Oncol 2015;33:1881-8. [Crossref] [PubMed]
- Jones PA, Issa JP, Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet 2016;17:630-41. [Crossref] [PubMed]
- Baumann M, Holscher T, Begg AC. Towards genetic prediction of radiation responses: ESTRO's GENEPI project. Radiother Oncol 2003;69:121-5. [Crossref] [PubMed]
- Thawani R, McLane M, Beig N, et al. Radiomics and radiogenomics in lung cancer: A review for the clinician. Lung Cancer 2018;115:34-41. [Crossref] [PubMed]
- Rutman AM, Kuo MD. Radiogenomics: creating a link between molecular diagnostics and diagnostic imaging. Eur J Radiol 2009;70:232-41. [Crossref] [PubMed]
- Segal E, Sirlin CB, Ooi C, et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol 2007;25:675-80. [Crossref] [PubMed]
- Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441-6. [Crossref] [PubMed]
- Chaddad A, Desrosiers C, Toews M. Phenotypic characterization of glioblastoma identified through shape descriptors. In: Medical Imaging: Computer-aided Diagnosis; 2016. 2016.
- Chaddad A, Desrosiers C, Toews M. GBM heterogeneity characterization by radiomic analysis of phenotype anatomical planes. In: Medical Imaging: Image Processing; 2016. 2016.
- Chaddad A, Tanougast C. High-throughput quantification of phenotype heterogeneity using statistical features. Adv Bioinformatics 2015;2015:728164. [Crossref] [PubMed]
- Yip SS, Aerts HJ. Applications and limitations of radiomics. Phys Med Biol 2016;61:R150-66. [Crossref] [PubMed]
- Limkin EJ, Sun R, Dercle L, et al. Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann Oncol 2017;28:1191-1206. [Crossref] [PubMed]
- Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology 2016;278:563-77. [Crossref] [PubMed]
- Chen B, Zhang R, Gan Y, et al. Development and clinical application of radiomics in lung cancer. Radiat Oncol 2017;12:154. [Crossref] [PubMed]
- Yoon HJ, Sohn I, Cho JH, et al. Decoding tumor phenotypes for ALK, ROS1, and RET fusions in lung adenocarcinoma using a radiomics approach. Medicine (Baltimore) 2015;94:e1753. [Crossref] [PubMed]
- Chen LD, Liang JY, Wu H, et al. Multiparametric radiomics improve prediction of lymph node metastasis of rectal cancer compared with conventional radiomics. Life Sci 2018;208:55-63. [Crossref] [PubMed]
- Yang X, Pan X, Liu H, et al. A new approach to predict lymph node metastasis in solid lung adenocarcinoma: a radiomics nomogram. J Thorac Dis 2018;10:S807-19. [Crossref] [PubMed]
- Zhong Y, Yuan M, Zhang T, et al. Radiomics approach to prediction of occult mediastinal lymph node metastasis of lung adenocarcinoma. AJR Am J Roentgenol 2018;211:109-13. [Crossref] [PubMed]
- Wu S, Zheng J, Li Y, et al. A Radiomics nomogram for the preoperative prediction of lymph node metastasis in bladder cancer. Clin Cancer Res 2017;23:6904-11. [Crossref] [PubMed]
- Zwanenburg A, Leger S, Vallières M, et al. Image biomarker standardisation initiative. In. p. arXiv preprint arXiv:1612.07003.
- Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. [Crossref] [PubMed]
- Zhao B, James LP, Moskowitz CS, et al. Evaluating variability in tumor measurements from same-day repeat CT scans of patients with non-small cell lung cancer. Radiology 2009;252:263-72. [Crossref] [PubMed]
- Coroller TP, Agrawal V, Narayan V, et al. Radiomic phenotype features predict pathological response in non-small cell lung cancer. Radiother Oncol 2016;119:480-6. [Crossref] [PubMed]
- Velazquez ER, Parmar C, Jermoumi M, et al. Volumetric CT-based segmentation of NSCLC using 3D-Slicer. Sci Rep 2013;3:3529. [Crossref] [PubMed]
- Johung KL, Yeh N, Desai NB, et al. Extended survival and prognostic factors for patients with ALK-rearranged non-small-cell lung cancer and brain metastasis. J Clin Oncol 2016;34:123-9. [Crossref] [PubMed]
- Cuneo KC, Nyati MK, Ray D, et al. EGFR targeted therapies and radiation: Optimizing efficacy by appropriate drug scheduling and patient selection. Pharmacol Ther 2015;154:67-77. [Crossref] [PubMed]
- Mak RH, Hermann G, Lewis JH, et al. Outcomes by tumor histology and KRAS mutation status after lung stereotactic body radiation therapy for early-stage non-small-cell lung cancer. Clin Lung Cancer 2015;16:24-32. [Crossref] [PubMed]
- Mak KS, Gainor JF, Niemierko A, et al. Significance of targeted therapy and genetic alterations in EGFR, ALK, or KRAS on survival in patients with non-small cell lung cancer treated with radiotherapy for brain metastases. Neuro Oncol 2015;17:296-302. [Crossref] [PubMed]
- Costa DB, Kobayashi S, Pandya SS, et al. CSF concentration of the anaplastic lymphoma kinase inhibitor crizotinib. J Clin Oncol 2011;29:e443-5. [Crossref] [PubMed]
- Dong Y, Feng Q, Yang W, et al. Preoperative prediction of sentinel lymph node metastasis in breast cancer based on radiomics of T2-weighted fat-suppression and diffusion-weighted MRI. Eur Radiol 2018;28:582-91. [Crossref] [PubMed]
- Tan X, Ma Z, Yan L, et al. Radiomics nomogram outperforms size criteria in discriminating lymph node metastasis in resectable esophageal squamous cell carcinoma. Eur Radiol 2019;29:392-400. [Crossref] [PubMed]
- Shen C, Liu Z, Wang Z, et al. Building CT radiomics based nomogram for preoperative esophageal cancer patients lymph node metastasis prediction. Transl Oncol 2018;11:815-24. [Crossref] [PubMed]
- Chaddad A, Desrosiers C, Toews M, et al. Predicting survival time of lung cancer patients using radiomic analysis. Oncotarget 2017;8:104393-407. [Crossref] [PubMed]
- Bianconi F, Fravolini ML, Bello-Cerezo R, et al. Evaluation of shape and textural features from CT as prognostic biomarkers in non-small cell lung cancer. Anticancer Res 2018;38:2155-60. [PubMed]
- Liu Y, Kim J, Balagurunathan Y, et al. Radiomic features are associated with EGFR mutation status in lung adenocarcinomas. Clin Lung Cancer 2016;17:441-448.e6. [Crossref] [PubMed]
- Coroller TP, Grossmann P, Hou Y, et al. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother Oncol 2015;114:345-50. [Crossref] [PubMed]
- Ferreira JJ, Koenigkam-Santos M, Cipriano F, et al. Radiomics-based features for pattern recognition of lung cancer histopathology and metastases. Comput Methods Programs Biomed 2018;159:23-30. [Crossref] [PubMed]


