Comparison of next-generation sequencing and immunohistochemistry analysis for targeted therapy-related genomic status in lung cancer patients
Introduction
The treatment of human cancer is shifting toward precision medicine, of which molecularly targeted therapy aimed at the genomic status of the tumor in each patient is a typic modality. Several drugs that target molecular pathways are available for patients with non-small cell lung cancer (NSCLC) harboring the relevant gene alterations.
Approximately 35% of NSCLC patients contain EGFR gene mutations (1), which are predictors of response to EGFR-tyrosine kinase inhibitors (TKIs) and predominantly located in exons 18 to 21 (2-6). In practical works, relevant mutations in patients are detected by direct sequencing (7), amplification refractory mutation system (ARMS) (7), digital PCR (dPCR) (8,9), next-generation sequencing (NGS), etc. (10,11). Immunohistochemistry (IHC) with mutation-specific antibodies (E746-A750Del and L858R) is also used to detect EGFR mutations and predict the response to EGFR-TKIs (12-15).
ALK/ROS1 inhibitors is another group of targeted drugs utilized mostly for the treatment for lung cancer patients with ALK- or ROS1-rearrangement (16,17). EML4-ALK fusion is present in approximately 4–6% of all NSCLC patients (18,19), and ROS1 rearrangements are present at an even lower frequency (1–2%) (18-20). In clinical application, the results of the ALK rearrangement by fluorescence in situ hybridization (FISH) and IHC are compared with each other to find the exact result (21). Positive staining of ROS1 by IHC should be double-checked by molecular assays to exclude false-positive cases because the specificity of IHC testing is not good enough (22). Massively parallel NGS assays are used in some clinical diagnostics to test for gene rearrangements (23,24).
In addition to the alterations of the three most common genes described above, some other lung cancer-related genes also play important roles. The EGFR pathway through RAS-RAF-MEK-MAPK and PI3K-AKT-mTOR can be activated by mutations in KRAS, NRAS, BRAF, ERBB2, RET, MET, FGFR1, or PIK3CA, resulting in resistance to TKIs (25-28), and TP53-inactivating mutations could disrupt its key function and are associated with poor prognosis (29).
If all the above molecular alterations need to be detected, a large amount of tumor tissue is necessary, with the exception of NGS. NGS has a considerable advantage in genetic mutation detection, but its sensitivity and accuracy of gene rearrangement detection are not clear. The aim of this study is to integrate the gene mutation and rearrangement alterations in a single targeted NGS panel to investigate the relevant gene alterations, and assess the accuracy of the NGS platform based on the integrated panel.
Methods
Patients and sampling
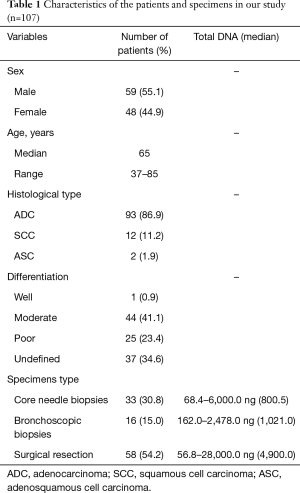
Formalin-fixed, paraffin-embedded (FFPE) tumor tissues from 107 NSCLC patients were collected from the Department of Pathology, Peking University First Hospital between April 1, 2018 and July 31, 2018. Characteristics of these patients were listed in Table 1. Thirty-three cases were core needle biopsies, 16 cases were bronchoscopic biopsies, and 58 cases were surgical resection specimens. All specimens were fixed in 10% neutral buffered formalin overnight. Sectioned tissues were routinely embedded in paraffin. For the patients with ALK or ROS1 fusion, response to the targeted drugs were recorded. The present study was approved by the Ethics Committee of Peking University First Hospital {No. 2016[1111]}.
Full table
IHC detection
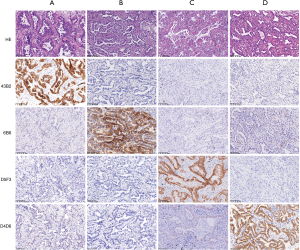
IHC staining tests were performed to explore the molecular status of EGFR (L858R and E746-A750del) on 65 samples, ALK rearrangement on 101 samples, and ROS1 rearrangement on 92 samples (Figure 1). IHC staining of tumor tissues was performed on 4-µm sections using the standard procedure and primary monoclonal antibodies against EGFR L858R (clone: 43B2, 1:200, Cell Signaling Technology), EGFR E746-A750del (clone: 6B6, 1:200, Cell Signaling Technology, Danvers, MA), ALK (clone: D5F3, 1:200, Ventana, Tucson, AZ), and ROS1 (clone: D4D6, 1:200, Cell Signaling Technology). The experiments were performed by standard protocols. A positive result was interpreted as moderate to strong staining of the membrane and/or cytoplasm in >10% tumor cells.
Nucleic acid extraction from tissue samples
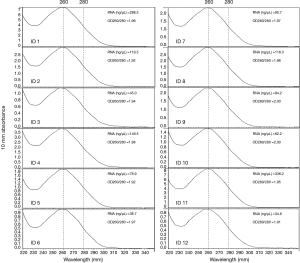
DNA was extracted from all the FFPE samples using the QIAamp DNA FFPE Tissue Kit (Qiagen, Hilton, Germany) according to the manufacturer’s instructions. The DNA was quantified using a Qubit Fluorometer 3.0 (Thermo Scientific, USA). A total mass of more than 20 ng and most fragments above 500 bp were suitable for the following NGS experiments. RNA was extracted from 12 cases with sufficient tissue using the RNeasy FFPE Kit (Qiagen, Hilton, Germany) according to the manufacturer’s instructions. The quantity and purity of the extracted RNA was measured using the NanoDrop ND-2000 Spectrophotometer (Thermo Scientific, MA, USA). A concentration higher than 20 ng/µL and an OD260/280 between 1.9 to 2.0 (Figure 2) is considered qualified for the quantitative PCR (qPCR) tests.
ARMS
ARMS PCR was performed on 40 cases using the AmoyDxTM Human EGFR Mutation Detection Kit (Amoy Diagnostics, Xiamen, China) for the detection of EGFR mutations in DNA sample, and was also performed on 12 RNA samples using the AmoyDxTM Human EML4-ALK/ROS1 Gene Fusions Detection Kit (Amoy Diagnostics) for the detection of ALK/ROS1 rearrangements. Each sample was detected with an external control assay and a mutation assay in the same well, while each run contained a negative control and a positive control. The amplification was set up according to the manufacturer’s instructions using the ABI 7500 real-time PCR system (Applied Biosystems, Foster City, CA). The final run files were also analyzed and interpreted according to the manufacturer’s instructions.
NGS platform and analysis pipeline
DNA library preparation and NGS sequencing for these cancer samples were performed according to the manufacturer’s recommended protocols of the SGI OncoAim® Lung Cancer Targeting Gene Detection Kit (Singlera Genomics Inc., Shanghai, China). Target regions were captured by designed probes targeting all exons of ten genes, including ALK, BRAF, ERBB2, EGFR, FGFR1, MET, KRAS, NRAS, PIK3CA and TP53, and potential gene rearrangement/fusion of ALK, ROS1 and RET.
The 150 bp paired-end sequencing was performed using a NextSeq 500 Sequencer (Illumina, Inc., San Diego, CA, USA) in combination with the NextSeq™ 500 High Output Kit (Illumina). Bioinformatics analysis of NGS sequence data was performed according to the guidelines of the OncoAim® Kit (Singlera), with read mapping, quality control, variant calling, and genotyping performed automatically using the Tools Kit supplied in the OncoAim® Kit (Singlera). Hg19/GRCh37 was used as the reference human genome sequence for aligning reads. Variant functional annotation was performed using the ENSEMBL Variant Effect Predictor tool. The minimum confidence threshold for variant and insertion/deletion (indel) calling was set to 5%.
Statistics
Comparisons of gene mutation frequency between adenocarcinoma (ADC) samples and squamous cell carcinoma (SCC) samples were performed using the chi-square test using SPSS v19.0 software. Cohen’s κ was calculated to assess the consistency of IHC staining and NGS. Cohen’s κ <0.4 was considered weak, 0.4≤ Cohen’s κ <0.75 was considered moderate, and Cohen’s κ ≥0.75 was considered strong. Significance was assumed for a P value of less than 0.05.
Results
DNA mutations in all patients
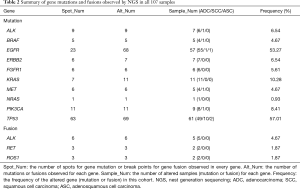
A total of 107 NSCLC patients, including 93 ADC, 12 SCC, and 2 adenosquamous carcinoma (ASC) cases, were enrolled in this study. DNA variations were found in 95 of the 107 carcinoma samples (88.79%). In total, 193 mutations in all ten genes and 12 gene rearrangements/fusions of all three genes in the NGS panel were observed (details of gene mutations and gene fusions detected by NGS in all 107 samples are available online: http://fp.amegroups.cn/cms/e5583e904e56e6a4d53175b2dbefdc56/jtd.2019.12.25-1.pdf). Overall, both the EGFR gene and the TP53 gene showed 69 mutations in our cases, which are much higher than that of other genes (Table 2). The hot spot mutation EGFR L858R and E746_A750del was found in 25 samples (23.4%) and 9 samples (8.4%) respectively, occurred with the two highest frequency. If we consider all types of exon 19 deletions of EGFR as one hot spot, we found 17 samples presented 5 types of EGFR 19Del, including c.2235_2249del (9 samples), c.2236_2250del (5 samples), c.2240_2257del (1 sample), c.2240_2254del (1 sample), c.2237_2253del (1 sample). Most EGFR mutations were found to be located in the EGFR kinase domain exon 19–21 in all patients. None or one mutation was observed in all SCC patients for the detected genes except TP53. A total of 11 mutations in the TP53 gene were found in ten SCC patients (10/12, 83.33%), which is more frequent than that in ADC patients (49/93, 52.69%, P=0.0440). The 12 gene fusions were all found in ADC samples (Table 2).
Full table
Real-time PCR (ARMS) confirmed the sensitivity of NGS
To assess the sensitivity and specificity of NGS for related gene alterations, we used ARMS to double check the gene status of EGFR for 40 cases, and ALK and ROS1 for 12 cases (Table 3). For one individual (ID 4), an EGFR G719C mutation found by NGS was not confirmed by ARMS. For patient ID 5 and ID 37, EGFR G719X mutations were observed by ARMS without distinguishing which nucleotide (adenine, thymine, or cytosine) replaced the guanine, whereas the results of NGS showed that guanine in 719 was changed into an adenine (G719A). Similarly, EGFR exon 20Ins was observed by ARMS in sample ID 6, and NGS discovered the inserted amino acid sequence and its position, which was shown as M766MASV. For other cases, the results of gene status of both mutations and fusions in the scope of the PCR kit detected by ARMS were completely consistent with that of NGS.

Full table
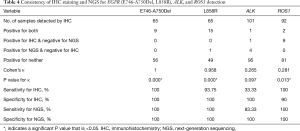
Concordance between NGS and IHC detection
High concordance between NGS and IHC on the detection of hotspots EGFR mutations
The results of IHC staining showed that 23.08% of samples were positive for EGFR L858R, of which 98.46% (64/65) samples showed consistent results between IHC staining and NGS. One sample was negative by IHC but positive by NGS, and this sample was confirmed to be positive for L858R by ARMS (sample ID 38). In the 65 samples, 9 samples showed either an c.2235_2249del or c.2236_2250del alteration, which were documented as positive for E746_A750del in the results of NGS. The same 9 samples were observed to be positive for E746_A750del by IHC staining as well, while all of the remaining 56 samples were negative for E746_A750del by either IHC or NGS, showing a high concordance between these two methods (Table 4 and Figure 3).
Full table
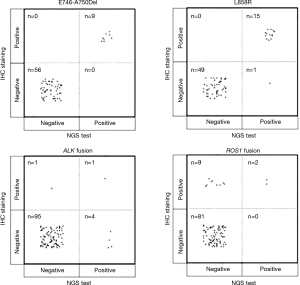
NGS could find more ALK fusion samples than IHC staining
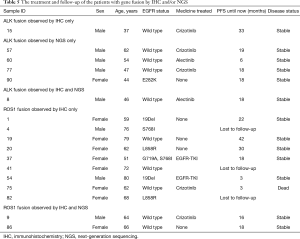
ALK rearrangement detection by IHC revealed two positive samples (2/101, 1.98%), whereas 5 positive samples were found by NGS (5/101, 4.95%). Among these ALK-positive samples, positive results by both IHC and NGS were shown in only one case (Figure 3), of which the disease was controlled stable after receiving therapy with alectinib (Table 5). For 5 of the 6 patients with ALK rearrangement revealed by either NGS or IHC, their tumors were all controlled stable or regressed after treatment with ALK inhibitors like crizotinib and alectinib (Table 5), indicating the facticity of all these results of ALK rearrangement. Based on this hypothesis, sensitivity (83.33%) of NGS was much higher than that (33.33%) of IHC (Table 4). The Cohen’s κ value of 0.265 suggested a low concordance between these two methods in ALK detection.
Full table
NGS could exclude the false positivity of ROS1 fusion detection by IHC
A total of 11 samples were positive for ROS1 rearrangement according to IHC staining (11/92, 11.96%), of which only two samples were demonstrated to be positive by NGS (2/92, 2.17%). These two samples were found stable of their disease, and one of them was receiving the treatment of crizotinib. Among the nine samples with ROS1 rearrangement by IHC only (Table 5), 6 were demonstrated to harbor EGFR mutations in exons 18 to 21 by NGS or ARMS PCR, 2 of which (patient ID 1 and 4) were demonstrated to be negative for ROS1 rearrangement by PCR amplification. The other three individuals, one was lost to follow-up, one still survived with a short follow-up time, and the last one progressed 3 months after the initial diagnosis, though treated with crizotinib, and died soon (Table 5). It was suggested that ROS1 rearrangement detected by IHC only were suspicious. Based on this hypothesis, a little lower specificity (90%) of IHC was observed than that (100%) of NGS (Table 4). The NGS results of ROS1 rearrangement were significantly different from the IHC results (Cohen’s κ 0.281, P=0.013).
Discussion
In this study, the NGS panel included not only the mutations found frequently in lung cancer but also 3 “druggable” fusion genes (ROS1, ALK, and RET) by a single NGS test with only 50 ng DNA. The genetic status in our study was in accordance with those in previous studies of NSCLC. For example, EGFR and TP53 mutated most frequently among these related genes (30,31), L858R and 19Del were the most common hot spots of the EGFR gene in NSCLC (6), and TP53 mutations were more prevalent in SCC patients than in ADC patients (31). Additionally, a high concordance was found between the results observed by ARMS and NGS, but more detailed information was revealed by NGS. All these items suggested the high reliability of the NGS experiments employed in our study.
For the results of EGFR L858R and E746-A750Del detected by NGS test, IHC staining showed almost the same results for these two molecular statuses. Previous studies (32,33) used two different rabbit monoclonal antibodies recognizing the EGFR mutations in exon 19 (E746-A750Del) and exon 21 (L858R) and showed a high sensitivity (91.5–93%) and specificity (100%) of IHC staining for L858R in comparison with direct sequencing. Our present study suggested that both IHC and NGS techniques are reliable for the clinical use of EGFR L858R detection. The unique type of DNA change for the mutation of L858R may contribute to the high consistency between IHC and direct or NGS sequencing. However, direct sequencing can find several types of DNA changes around the EGFR E746-A750 region, some of which could give different amino changes when detected with the antibodies used in IHC staining, resulting in a lower sensitivity (63–77%) of E746-A750Del by IHC compared with direct sequencing in the previous studies (32,33) mentioned above. According to the NGS results, we could distinguish the different types of alterations in this region. We could also easily detect the exacted DNA changes occurring in the E746-A750Del, which could be observed by IHC staining with the mutation-specific antibody, and improved the consistency of IHC and NGS to 100% in our study. At the same time, more information, including single nucleotide variants (SNVs) in exons 18–21 and other types of EGFR 19Del, could be determined by NGS, thereby predicting the response to the EGFR-TKIs more reliably, rather than IHC staining (34).
The results of the ALK fusion detection by NGS showed an acceptable frequency of 4.7%, which was similar to the reported rate of 4–8% (18,19). Detection of ALK rearrangement by protein-based IHC staining usually showed a higher sensitivity than other methods, such as DNA-based FISH and mRNA-based RT-PCR (35). The criteria for FISH positive results are artificial and depend too much on the subjectivity of the pathologist (18). The RT-PCR method relies on primers that cannot consider all types of ALK fusion. However, any effective fusion will result in the fusion protein, which can be observed by IHC staining. NGS shows a higher sensitivity than FISH in detecting ALK rearrangements (36). However, no matter whether IHC staining, NGS, FISH or RT-PCR was used, it cannot achieve an accuracy rate of 100% on the ALK rearrangement detection (37). In our study, one sample was ALK positive for IHC staining but negative for NGS, of which the break point of ALK in this sample may be outside of the panel used in the NGS test. And this ALK fusion was confirmed to be true positive by the response of the patient to the targeted therapy. In contrast, ALK fusion identified by NGS but negative by IHC staining were found in four individuals, most of which the reality of these gene fusion was confirmed by their responses to the targeted treatment. Based on these results, ALK positive by either IHC or NGS deserved serious consideration. While in the situation of ALK negative identified by IHC or NGS only, more targeted methods like FISH and RT-PCR should be performed to avoid missing the potential opportunity to benefit from the targeted drugs.
In our study, 11 samples (11/92, 11.96%) were positive for ROS1 by IHC staining, of which 9 samples were ROS1 negative by NGS. The positive rate of ROS1 by IHC staining in our study was unrealistically higher than that detected by NGS (1.87%), as well as that reported in existing publications (1–2%) (18-20). It was extremely rare that patients harbor both EGFR mutation and ROS1 fusion. There was only one reported case that simultaneous L858R mutation and ROS1 fusion occurred in a single NSCLC patient with intrinsic gefitinib resistance (38). In our cohort, more than a half of samples with ROS1 fusion identified by IHC only showed simultaneous EGFR mutation by NGS or ARMS PCR, 2 of whom responded to the EGFR-TKI, which made the IHC results extremely suspicious. In previously published studies (18,39), ROS1 positive rates were commonly presented with different results when multiple methods were used in one cohort, and it has been suggested that IHC was not sensitive enough to determine ROS1 rearrangement. ROS1 staining could be observed in certain nonneoplastic conditions, including normal pneumocytes and bronchiolar metaplasia, which might result in false-positive staining for ROS1 rearrangement detection by IHC staining (40,41). Our results were in concordance with the conclusion that the sensitivity of ROS1 detection by IHC staining is much better than its specificity (22,39). IHC is an effective screening method for avoiding other redundant analyses in patients with ROS1 negative results (39). However, for ROS1-positive samples detected by IHC staining, it is necessary to perform other analyses, such as FISH or NGS, for further validation.
The mutation frequencies of the remaining genes detected by NGS was in accordance with those in reported studies of NSCLC, such as KRAS (8.93–30%), MET (5%), ERBB2 (1.79–4%), FGFR1 (1–3%), NRAS (<2%), PIK3CA (5.36%) and BRAF (1.79–6%) (30). These results suggest that NGS could comprehensively investigate the status of many lung cancer-related genes in addition to popular druggable genes (EGFR, ALK, and ROS1).
Conclusions
Various gene mutations and gene rearrangements could be detected by a single platform of NGS, which showed high performance and cost-effectiveness. NGS could provide more informative and reliable results than IHC staining for EGFR gene alterations, especially for the exon 19 region. For gene fusion detection, NGS increased the positive rate of ALK and decreased the false-positive results of ROS1 compared with IHC staining. Thus, we suggest that in the samples with ALK negative and/or ROS1 positive observed by IHC only, it is necessary to perform other analyses like NGS for confirmation.
Acknowledgments
We thank all researchers for their contributions in this study, including those whose relevant contributions were not cited due to space limitations.
Funding: This work was supported by Beijing Municipal Science and Technology Commission (No. Z181100001918007).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethics Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Ethics Committee of Peking University First Hospital {No. 2016[1111]}. Ethics approval and informed consent to participate were given by all patients in writing.
References
- Rossi G, Graziano P, Leone A, et al. The role of molecular analyses in the diagnosis and treatment of non-small-cell lung carcinomas. Semin Diagn Pathol 2013;30:298-312. [Crossref] [PubMed]
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. [Crossref] [PubMed]
- Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A 2004;101:13306-11. [Crossref] [PubMed]
- Greig SL. Osimertinib: First Global Approval. Drugs 2016;76:263-73. [Crossref] [PubMed]
- Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst 2005;97:339-46. [Crossref] [PubMed]
- Borras E, Jurado I, Hernan I, et al. Clinical pharmacogenomic testing of KRAS, BRAF and EGFR mutations by high resolution melting analysis and ultra-deep pyrosequencing. BMC Cancer 2011;11:406. [Crossref] [PubMed]
- Ellison G, Donald E, McWalter G, et al. A comparison of ARMS and DNA sequencing for mutation analysis in clinical biopsy samples. J Exp Clin Cancer Res 2010;29:132. [Crossref] [PubMed]
- Malapelle U, de Luca C, Vigliar E, et al. EGFR mutation detection on routine cytological smears of non-small cell lung cancer by digital PCR: a validation study. J Clin Pathol 2016;69:454-7. [Crossref] [PubMed]
- Wang J, Ramakrishnan R, Tang Z, et al. Quantifying EGFR alterations in the lung cancer genome with nanofluidic digital PCR arrays. Clin Chem 2010;56:623-32. [Crossref] [PubMed]
- Tuononen K, Mäki-Nevala S, Sarhadi VK, et al. Comparison of targeted next-generation sequencing (NGS) and real-time PCR in the detection of EGFR, KRAS, and BRAF mutations on formalin-fixed, paraffin-embedded tumor material of non-small cell lung carcinoma-superiority of NGS. Genes Chromosomes Cancer 2013;52:503-11. [Crossref] [PubMed]
- Lin MT, Mosier SL, Thiess M, et al. Clinical validation of KRAS, BRAF, and EGFR mutation detection using next-generation sequencing. Am J Clin Pathol 2014;141:856-66. [Crossref] [PubMed]
- Kitamura A, Hosoda W, Sasaki E, et al. Immunohistochemical detection of EGFR mutation using mutation-specific antibodies in lung cancer. Clin Cancer Res 2010;16:3349-55. [Crossref] [PubMed]
- Azuma K, Okamoto I, Kawahara A, et al. Association of the expression of mutant epidermal growth factor receptor protein as determined with mutation-specific antibodies in non-small cell lung cancer with progression-free survival after gefitinib treatment. J Thorac Oncol 2012;7:122-7. [Crossref] [PubMed]
- Yu J, Kane S, Wu J, et al. Mutation-specific antibodies for the detection of EGFR mutations in non-small-cell lung cancer. Clin Cancer Res 2009;15:3023-8. [Crossref] [PubMed]
- Xiong Y, Bai Y, Leong N, et al. Immunohistochemical detection of mutations in the epidermal growth factor receptor gene in lung adenocarcinomas using mutation-specific antibodies. Diagn Pathol 2013;8:27. [Crossref] [PubMed]
- Solomon BJ, Mok T, Kim DW, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med 2014;371:2167-77. [Crossref] [PubMed]
- Shaw AT, Ou SH, Bang YJ, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med 2014;371:1963-71. [Crossref] [PubMed]
- Reguart N, Teixido C, Gimenez-Capitan A, et al. Identification of ALK, ROS1, and RET Fusions by a Multiplexed mRNA-Based Assay in Formalin-Fixed, Paraffin-Embedded Samples from Advanced Non-Small-Cell Lung Cancer Patients. Clin Chem 2017;63:751-60. [Crossref] [PubMed]
- Takeuchi K, Soda M, Togashi Y, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med 2012;18:378-81. [Crossref] [PubMed]
- Gainor JF, Shaw AT. Novel targets in non-small cell lung cancer: ROS1 and RET fusions. Oncologist 2013;18:865-75. [Crossref] [PubMed]
- Murakami Y, Mitsudomi T, Yatabe Y. A Screening Method for the ALK Fusion Gene in NSCLC. Front Oncol 2012;2:24. [Crossref] [PubMed]
- Su Y, Goncalves T, Dias-Santagata D, et al. Immunohistochemical Detection of ROS1 Fusion. Am J Clin Pathol 2017;147:77-82. [PubMed]
- Davies KD, Le AT, Sheren J, et al. Comparison of Molecular Testing Modalities for Detection of ROS1 Rearrangements in a Cohort of Positive Patient Samples. J Thorac Oncol 2018;13:1474-82. [Crossref] [PubMed]
- Rossi G, Ragazzi M, Tamagnini I, et al. Does Immunohistochemistry Represent a Robust Alternative Technique in Determining Drugable Predictive Gene Alterations in Non-Small Cell Lung Cancer? Curr Drug Targets 2017;18:13-26. [Crossref] [PubMed]
- Ai X, Guo X, Wang J, et al. Targeted therapies for advanced non-small cell lung cancer. Oncotarget 2018;9:37589-607. [Crossref] [PubMed]
- Nelson-Taylor SK, Le AT, Yoo M, et al. Resistance to RET-Inhibition in RET-Rearranged NSCLC Is Mediated By Reactivation of RAS/MAPK Signaling. Mol Cancer Ther 2017;16:1623-33. [Crossref] [PubMed]
- Bui KT, Cooper WA, Kao S, et al. Targeted Molecular Treatments in Non-Small Cell Lung Cancer: A Clinical Guide for Oncologists. J Clin Med 2018. [Crossref] [PubMed]
- Tetsu O, Hangauer MJ, Phuchareon J, et al. Drug Resistance to EGFR Inhibitors in Lung Cancer. Chemotherapy 2016;61:223-35. [Crossref] [PubMed]
- Shepherd FA, Lacas B, Le Teuff G, et al. Pooled Analysis of the Prognostic and Predictive Effects of TP53 Comutation Status Combined With KRAS or EGFR Mutation in Early-Stage Resected Non-Small-Cell Lung Cancer in Four Trials of Adjuvant Chemotherapy. J Clin Oncol 2017;35:2018-27. [Crossref] [PubMed]
- Jing C, Mao X, Wang Z, et al. Next-generation sequencing-based detection of EGFR, KRAS, BRAF, NRAS, PIK3CA, Her2 and TP53 mutations in patients with non-small cell lung cancer. Mol Med Rep 2018;18:2191-7. [PubMed]
- Mogi A, Kuwano H. TP53 mutations in nonsmall cell lung cancer. J Biomed Biotechnol 2011;2011:583929. [Crossref] [PubMed]
- Kawahara A, Azuma K, Sumi A, et al. Identification of non-small-cell lung cancer with activating EGFR mutations in malignant effusion and cerebrospinal fluid: rapid and sensitive detection of exon 19 deletion E746-A750 and exon 21 L858R mutation by immunocytochemistry. Lung Cancer 2011;74:35-40. [Crossref] [PubMed]
- Simonetti S, Molina MA, Queralt C, et al. Detection of EGFR mutations with mutation-specific antibodies in stage IV non-small-cell lung cancer. J Transl Med 2010;8:135. [Crossref] [PubMed]
- Wu M, Pan X, Xu Y, et al. Methodological comparison of the allele refractory mutation system and direct sequencing for detecting EGFR mutations in NSCLC, and the association of EGFR mutations with patient characteristics. Oncol Lett 2018;16:1087-94. [PubMed]
- Wynes MW, Sholl LM, Dietel M, et al. An international interpretation study using the ALK IHC antibody D5F3 and a sensitive detection kit demonstrates high concordance between ALK IHC and ALK FISH and between evaluators. J Thorac Oncol 2014;9:631-8. [Crossref] [PubMed]
- Ali SM, Hensing T, Schrock AB, et al. Comprehensive Genomic Profiling Identifies a Subset of Crizotinib-Responsive ALK-Rearranged Non-Small Cell Lung Cancer Not Detected by Fluorescence In Situ Hybridization. Oncologist 2016;21:762-70. [Crossref] [PubMed]
- Letovanec I, Finn S, Zygoura P, et al. Evaluation of NGS and RT-PCR Methods for ALK Rearrangement in European NSCLC Patients: Results from the European Thoracic Oncology Platform Lungscape Project. J Thorac Oncol 2018;13:413-25. [Crossref] [PubMed]
- Qiu D, Zhang Y, Xue YB, et al. Chemotherapy combined with Endostar as salvage treatment for EGFR-tyrosine kinase inhibitor primary resistance in an advanced non-small cell lung cancer patient with EGFR L858R mutation and ROS1 fusion: A case report. Thorac Cancer 2019;10:1023-8. [Crossref] [PubMed]
- Rossi G, Jocolle G, Conti A, et al. Detection of ROS1 rearrangement in non-small cell lung cancer: current and future perspectives. Lung Cancer (Auckl) 2017;8:45-55. [Crossref] [PubMed]
- Sholl LM, Sun H, Butaney M, et al. ROS1 immunohistochemistry for detection of ROS1-rearranged lung adenocarcinomas. Am J Surg Pathol 2013;37:1441-9. [Crossref] [PubMed]
- Cha YJ, Lee JS, Kim HR, et al. Screening of ROS1 rearrangements in lung adenocarcinoma by immunohistochemistry and comparison with ALK rearrangements. PLoS One 2014;9:e103333. [Crossref] [PubMed]