
Frequency of MET exon 14 skipping mutations in non-small cell lung cancer according to technical approach in routine diagnosis: results from a real-life cohort of 2,369 patients
Introduction
Oncogenic alterations occurring in non-small cell lung cancer (NSCLC) are attractive because they may confer sensitivity to targeted therapies and help personalise medicine with the use of targeted therapy (1). Alterations of the gene coding for the mesenchymal epithelial transition receptor (MET), a tyrosine kinase receptor, have been reported in NSCLC, including copy number variations (polysomy and amplification) and mutational activating variants (2,3). According to the Cancer Genome Atlas, the latter occur in 7% of pulmonary adenocarcinoma (2) with different frequencies according to ethnicity (4,5). The most frequent mutations are MET exon 14 skipping mutations that activate MET through reduction of MET degradation and increased MET activity (6,7). Because of the highly diverse sequence composition of MET exon 14 skipping mutations, the frequency of these mutations is strongly dependent on the technique used to detect MET alterations. It varies from 3% of pulmonary adenocarcinoma using direct sequencing or hybrid capture new generation sequencing (7,8) to 4.3% using whole-exome sequencing (2).
The purpose of this retrospective study was to describe the frequency of MET exon 14 skipping mutations according to the different technical changes that have been used over recent years in a real-life context.
Methods
Patients and tumour samples
Between March 2014 and May 2018, tissue samples from patients with metastatic NSCLC were analysed for MET exon 14 skipping mutation as part of routine practice in the Pathology Department of the Hospices Civils de Lyon, France. A consent form was systematically obtained through the umbrella protocol of the biological specimen resource core facility of the Hospices Civils de Lyon. The present study was approved by the ethics committee of the Hospices Civils de Lyon. Clinical data, such as gender, age, smoking history, stage of the disease, and histopathological reports were retrospectively collected from all centres which had referred the cases for molecular diagnosis and ultimately managed the patients. Based on medical histopathological report and/or histopathological slides reviewed by two thoracic pathologists, lung carcinomas were classified according to the World Health Organization histopathological classification (1).
C-MET protein expression
Standard 4-µm formalin-fixed paraffin-embedded (FFPE) sections were subjected to immunohistochemical (IHC) analysis using a c-MET antibody (SP44, Ventana, Basel, Switzerland). Staining for c-MET was scored semi-quantitatively on a four-tier scale from 0 (absent) to 3 (strong membranous staining in >50% of tumour cells) as previously described (9). In case of a score of 2 (moderate expression) or 3 (high expression), fluorescent in situ hybridization (FISH) analysis was performed to confirm MET amplification.
MET amplification
MET gene copy number amplification was assessed by FISH using the Zytolight, MET/CEN7 dual color probe kit (Clinisciences, Nanterre, France). MET amplification was defined as a gene copy number ≥6 in at least 100 tumour cell nuclei or as the presence of clusters in >3 nuclei. Polysomy was defined as a ratio MET:CEN7 <2. In cases of exhausted samples, the analysis was performed on at least 25 cells.
MET gene exon 14 analysis
DNA was extracted from FFPE lung tumour tissue after laser microdissection (Leica, LMD 6000, Wetzlar, Germany; QIAmp DNA micro-kit, QIAGEN, Venlo, The Netherlands). In the Hospices Civils de Lyon, routine MET exon 14 genetic alterations were detected by three different methods over the duration of the study: (I) from 1 March 2014 to 30 April 2016 (period 1), Sanger sequencing of MET exon 14 and its splice sites (NM 001127500.2: c.2942-106 to c.3082+104) was performed only upon clinician request after investigation of other drivers by multiplex KRAS/EGFR/BRAF Sanger sequencing (KRAS/EGFR/BRAF negative + MET Sanger sequencing); (II) from 1 May 2016 to 31 August 2017 (period 2), next generation sequencing (NGS) was performed systematically for prospective samples using a custom ampliseq panel including the part of the MET exon 14 coding region (NM 001127500.2:c.2942-5 to c.3082+54) and Ion Personal Genome Machine technology (PGM Ion Torrent, ThermoFisher Scientific, Waltham, MA, USA) (period 2, NGS panel V1 only). Since this coverage is insufficient for MET splice site mutation detection (in particular at the 5’ end) and in case wild-type KRAS/EGFR/BRAF, Sanger sequencing completed the analysis upon clinician request (period 2, NGS panel V1 negative + MET Sanger sequencing). For some previous samples, already analysed for the main oncogenic drivers before the beginning of the second period, only Sanger sequencing was performed upon clinician request (period 2, KRAS/EGFR/BRAF negative + MET Sanger sequencing); (III) from 1 September 2017 to 30 May 2018 (period 3), the custom ampliseq panel was modified to cover MET exon 14 splice sites (NM 001127500.2: c.2942-97 to c.2994 and c.3031 to c.3082+181; NGS V2). In case of unpublished MET exon 14 mutations, MET exon 14 skipping was confirmed by an RNA-approach technique currently available in our department. The interested reader can find details about the technical aspects of NGS and RNA assay in the Supplementary appendix online.
The MET exon 14 molecular analysis was performed after (period 1) or in parallel (period 2 and 3) of EGFR/KARS/BRAF/HER2, screened by sequencing and of ALK and ROS1, screened by immunohistochemistry.
Results
Patients and samples characteristics
A total of 2,369 samples from patients with NSCLC were analyzed for MET exon 14 skipping mutations from March 2014 until May 2018. Among these, 62 (2.6%) tumours harboured MET exon 14 genetic alterations. Patients were mainly women, (61% of cases) and the median age was 75 years (range, 46–97 years). Clinical data are summarized in Table 1.

Full table
Histopathological characteristics
Among the 62 samples harbouring a MET exon 14 mutation, 47 assays were performed on small samples (endoscopic/percutaneous biopsies or cell-block, 76%). Among these 47 assays, 38 (81%) were taken from bronchial biopsies, 7 (15%) from percutaneous biopsies and two were cytological samples (4%). In addition, 15/62 (24%) were taken from lung or pleural surgical biopsies. Overall, 49 (79%) tumours were adenocarcinomas, 5 (8%) were squamous carcinomas, 4 (6%) were sarcomatoid carcinomas, and 4 (6%) were classified as NSCLC-NOS (not otherwise specified; Table 1). MET IHC analysis was performed on 22 samples; 19 (86%) of these were positive (Figure S1). FISH analysis, performed on positive cases, found amplification (n=2; Figure S2) or polysomy (n=1) in 3/19 (16%) cases (Table S1).
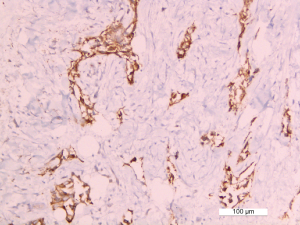
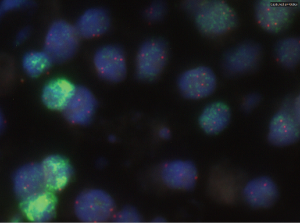
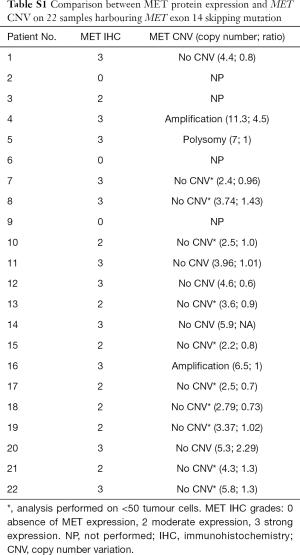
Full table
Mutational characteristics
Considering all study periods, genomic alterations on the skipping site of MET exon 14 were detected in 2.6% of lung NSCLC. The description of the mutation type and its implication on the donor or acceptor site are described in Figure 1. Among these 62 mutations, 50 (81%) cases were diagnosed by NGS, while 12 (19%) mutations were identified by Sanger sequencing. During the second period one mutation in the part of the gene not covered by the NGS panel V1was diagnosed by Sanger sequencing (Table 2). The frequency of MET exon 14 alterations varied depending on the selection of patients and technical approach used; the highest was found in patients who were known to have wild-type KRAS/EGFR/BRAF (15.4%). Among patients in whom only NGS was used, the frequency of MET exon 14 alterations was the greatest when the NGS panel covered the 3’ and 5’ of MET exon 14 (panel V2, 4.1%; Table 3). Co-occurring genomic mutations were present in three cases (5%) including one KRAS G12C mutation, one NRAS Q61L mutation and one MET H1112Y substitution.
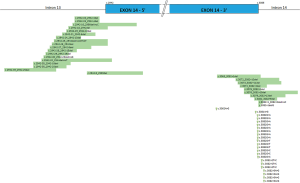

Full table

Full table
Discussion
In this large cohort of patients with NSCLC, we show that the frequency of MET exon 14 skipping mutations differed over time in accordance with the technical approach used. Other than clinical correlates, we also report the co-occurrence of RAS and MET mutations.
The present study confirms some clinical and histopathological data. First, MET exon 14 skipping mutation were mostly observed in women and those who had never smoked, which is in line with previously reported features of patients with NSCLC harbouring oncogenic driver alteration (10); interestingly, the median age of the population was higher than that reported in case of EGFR or ALK alteration (4,11). This has a significant potential clinical relevance as elderly people are often not able to be treated by classical chemotherapy and targeted therapy could be a less toxic alternative. Furthermore, MET exon 14 skipping mutations were first described in sarcomatoid carcinoma (12-14), but herein they were mainly diagnosed in lung adenocarcinoma, as has widely been reported elsewhere (7,15,16). This underlines that the molecular screening of such mutations should not be limited to sarcomatoid carcinoma. The study also confirms that MET immunohistochemistry does not represent a valuable tool to screen patients for MET genomic alterations, including MET exon 14 skipping mutation as well as MET amplification. Furthermore, the results herein also confirm that co-occurring mutations are observed in 5% of NSCLC specimens that harbour a MET exon 14 skipping alteration (5,12,17) which precludes the prescription of MET tyrosine kinase inhibitors for such patients. Ultimately, a key clinical point for routine practice is that screening for MET exon 14 skipping mutations should be systematic irrespective of the tumour status for other oncogenic driver alteration, and the clinical characteristics of patients.
The main purpose of this study was to analyse the impact of technical approach to detect MET exon 14 skipping mutations in routine practice. For instance, there were differences in the frequency of MET exon 14 skipping mutations according to the period of study and the sequencing design. When MET alterations were detected by Sanger sequencing in patients known to have wild-type KRAS/EGFR/BRAF, the frequency was very high which is explained mainly by the selection of the patients. To better evaluate this frequency, it would have been interesting to consider all patients who had received molecular analysis, but this was not possible to identify such individuals for the first study period. The lowest frequency of MET exon 14 skipping mutations detection was observed during the period when NGS was used systematically but the panel only covered part of the exon 14 and without further Sanger sequencing. In the most recent period, patients were not selected and the NGS panel covered the 3’ and 5’ of MET exon 14; the frequency of MET exons 14 skipping mutations is the same as that reported in other studies, including those using whole-exome sequencing (2) and a RNA-based assay (18).
Furthermore, three-quarters of the MET alterations were diagnosed on small samples in line with routine practice in the setting of advanced and metastatic disease and FFPE specimens. Recently, Davies et al. (18) described the higher sensitivity of the RNA-based NGS assay employing anchored multiplex PCR for target enrichment comparing to a DNA-based NGS assay that employs an amplicon-mediated target enrichment, the latter being the approach used herein. Furthermore, RNA-based NGS assay allow the detection of ALK, ROS1, RET, NTRK1, and NRG1 translocations in a single assay, that could be suitable for small samples. However, as expected, some samples herein that were positive using the DNA-based approach were uninformative when analysed by the RNA-based approach due to the poor quality or low quantity of RNA. In routine practice, we do not often have the possibility to work on frozen samples or on a large amount of tissue and the assay used should be adapted to these aspects. To perform biomarker analysis on all kinds of samples, including cytological samples, a DNA-based NGS assay that employs an amplicon-mediated target enrichment is a more suitable technique to detect the more frequent genomic alterations. The frequency of MET exon 14 skipping mutations observed in the most recent period of this study confirm that a DNA-based assay with a good coverage of MET exon 14 skipping mutations could detect these alterations with a same frequency as an RNA-based assay in an unselected population.
To conclude, the frequency of genomic alterations is highly dependent on patient selection and the technical approach. For routine practice, a compromise has to be made between sensitivity and feasibility, the DNA-based NGS assay used herein seems to be a practical alternative to RNA-based techniques for small FFPE specimens.
Supplementary
Sequence analysis
DNA was extracted from FFPE lung tumour tissue after laser microdissection ensuring a high (>60%) tumour content (Leica, LMD 6000, QIAmp DNA micro-kit, QIAGEN, Venlo, The Netherlands). DNA was quantified with a Qubit fluorometer (Thermofischer, Waltham, MA, USA). PCR and sequencing reactions were performed in duplicate using a 1 to 5 ng DNA input. Regions sequenced around MET exon 14 are specified in the main text. Samples were defined as wild-type only in the absence of variants with a minimal depth of 300×. Only variants present on both replicas were considered as mutations.
mRNA analysis
RNA was extracted from FFPE lung tumour tissue after macrodissection ensuring a least a 20% tumour content (RNeasy FFPE kit, QIAGEN). RNA was quantified with a Qubit fluorometer (Thermofischer) and 25 to 200 ng RNA were hybridized with a Nanostring (Seattle, WA, USA) custom panel comprising probes for MET exon 13/14 junction and for MET exon 13/15 junction. Count were recorded by a nCounter Digital Analyser (Nanostring). RNA quality and quantity were checked using four housekeeping genes: counts should >100 for GUSB and POLR2A and over 1,000 for GAPDH and OAZ1. Counts >100 for the exon 13/15 junction indicate a MET exon 14 skipping.
Acknowledgments
The authors thank Dominique Bourchany, Arlette Loiseau, and Pauline Desormeaux (Lyon university hospital, France) for performing MET immunohistochemistry and MET FISH analysis. We thank Pr Antoinette Lemoine (Paul Brousse Hospital, Paris) and Dr Philip Robinson (DRCI, Hospices Civils de Lyon) for help in manuscript preparation. The authors also thank the pathologists from the Rhône-Alpes and the Biological Resource Center of the Hospices Civils de Lyon (Tissu-tumorothèque Est).
Funding: This work was supported by the French Ministries for Health and Research (lyric grant INCa-DGOS-4664), the French National Cancer Institute (INCa) and by the ARISTOTE association (AS).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd.2020.04.21). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The present study was approved by the ethics committee of the Hospices Civils de Lyon (No. N° 19-17). A consent form was systematically obtained through the umbrella protocol of the biological specimen resource core facility of the Hospices Civils de Lyon.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol 2015;10:1243-60. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. Erratum in: Nature. 2014 Oct 9;514(7521):262. Rogers, K [corrected to Rodgers, K]. Author Correction: Comprehensive molecular profiling of lung adenocarcinoma. [Nature. 2018]. [Crossref] [PubMed]
- Salgia R. MET in Lung Cancer: Biomarker Selection Based on Scientific Rationale. Mol Cancer Ther 2017;16:555-65. [Crossref] [PubMed]
- Lee GD, Lee SE, Oh DY, et al. MET Exon 14 Skipping Mutations in Lung Adenocarcinoma: Clinicopathologic Implications and Prognostic Values. J Thorac Oncol 2017;12:1233-46. [Crossref] [PubMed]
- Schrock AB, Frampton GM, Suh J, et al. Characterization of 298 Patients with Lung Cancer Harboring MET Exon 14 Skipping Alterations. J Thorac Oncol 2016;11:1493-502. [Crossref] [PubMed]
- Cortot AB, Kherrouche Z, Descarpentries C, et al. Exon 14 Deleted MET Receptor as a New Biomarker and Target in Cancers. J Natl Cancer Inst 2017. [Crossref] [PubMed]
- Frampton GM, Ali SM, Rosenzweig M, et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov 2015;5:850-9. [Crossref] [PubMed]
- Onozato R, Kosaka T, Kuwano H, et al. Activation of MET by gene amplification or by splice mutations deleting the juxtamembrane domain in primary resected lung cancers. J Thorac Oncol 2009;4:5-11. [Crossref] [PubMed]
- Castiglione R, Alidousty C, Holz B, et al. Comparison of the genomic background of MET-altered carcinomas of the lung: biological differences and analogies. Mod Pathol 2019;32:627-38. [Crossref] [PubMed]
- Zheng D, Wang R, Ye T, et al. MET exon 14 skipping defines a unique molecular class of non-small cell lung cancer. Oncotarget 2016;7:41691-702. [Crossref] [PubMed]
- Awad MM, Oxnard GR, Jackman DM, et al. MET Exon 14 Mutations in Non-Small-Cell Lung Cancer Are Associated With Advanced Age and Stage-Dependent MET Genomic Amplification and c-Met Overexpression. J Clin Oncol 2016;34:721-30. [Crossref] [PubMed]
- Saffroy R, Fallet V, Girard N, et al. MET exon 14 mutations as targets in routine molecular analysis of primary sarcomatoid carcinoma of the lung. Oncotarget 2017;8:42428-37. [Crossref] [PubMed]
- Wislez M, Domblides C, Cortot A, et al. Mutations at the splice sites of exon 14 of MET gene: a new target for sarcomatoid carcinomas?. Ann Transl Med 2016;4:96. [Crossref] [PubMed]
- Liu X, Jia Y, Stoopler MB, et al. Next-Generation Sequencing of Pulmonary Sarcomatoid Carcinoma Reveals High Frequency of Actionable MET Gene Mutations. J Clin Oncol 2016;34:794-802. [Crossref] [PubMed]
- Paik PK, Drilon A, Fan PD, et al. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. Cancer Discov 2015;5:842-9. Erratum in: Cancer Discov. 2016 Mar;6(3):330. [Crossref] [PubMed]
- Kim EK, Kim KA, Lee CY, et al. Molecular Diagnostic Assays and Clinicopathologic Implications of MET Exon 14 Skipping Mutation in Non-small-cell Lung Cancer. Clin Lung Cancer 2019;20:e123-32. [Crossref] [PubMed]
- Bruno R, Lupi C, Landi L, et al. MET exon 14 mutations in advanced lung adenocarcinoma: Frequency and coexisting alterations. J Clin Oncol 2017;35:e20656. [Crossref]
- Davies KD, Lomboy A, Lawrence CA, et al. DNA-Based versus RNA-Based Detection of MET Exon 14 Skipping Events in Lung Cancer. J Thorac Oncol 2019;14:737-41. [Crossref] [PubMed]


