
MiR-212-3p mediates apoptosis and invasion of esophageal squamous cell carcinoma through inhibition of the Wnt/β-catenin signaling pathway by targeting SOX4
Introduction
Esophageal squamous cell carcinoma (ESCC) is among the most common malignancies in the world. In recent years, the incidence of ESCC has risen gradually, and it is ranked eighth and sixth out of all cancers for incidence and mortality, respectively (1,2). ESCC severely impacts the health and life of patients, but its exact pathogenesis has yet to be fully understood. Despite recent technological improvements in the diagnosis and treatment of ESCC, no obvious decrease has been observed in the mortality rate (3). Surgery, radiotherapy, and chemotherapy constitute the main approaches for treating ESCC. Due to the insidious onset and atypical symptoms of early ESCC, effective clinical methods for early diagnosis are lacking. Moreover, the esophageal submucosa has an abundant lymphatic capillary network. As a result, 70–80% of ESCC patients already have lymph node and distant metastasis at the time of diagnosis. For patients with advanced ESCC, drug therapy is still the main treatment; however, the general efficacy is unsatisfactory (4,5). Therefore, it is especially important that the molecular mechanisms underlying the occurrence, development, and metastasis of ESCC are urgently explored, so that prevention and treatment targets can be developed and new intervention strategies for ESCC can be established.
Gene intervention treatment is a research hotpot in cancer prevention and treatment. The role of microRNAs (miRNAs) has attracted an increasing amount of attention in studies on the regulation of oncogene expression. miRNAs are small, endogenous, single-stranded non-coding RNAs in eukaryotic organisms. MiRNAs were recently discovered to play an important role in cell proliferation, differentiation, and apoptosis, as well as the regulation of eukaryotic gene expression. MiRNA molecules bind to the 3' untranslated region (UTR) of target genes by forming an RNA-induced silencing complex (RISC) in an incompletely complementary way and inhibit their expression by inducing mRNA cleavage, degradation, translational repression, or other regulatory mechanisms. The importance of miRNA dysregulation in the occurrence, development, invasion, and metastasis of ESCC has been confirmed. For instance, miR-183 promotes ESCC proliferation and invasion by targeting programmed cell death (6), while miR-503 promotes tumor progression and serves as a novel prognostic biomarker in ESCC (7). Recently, studies have shown that abnormal expression of miR-212-3p has been found in many tumors, suggesting that it may be related to tumor development. Some studies have demonstrated that miR-212-3p expression is down-regulated in oral squamous cells (8). Therefore, miR-212-3p may play an important part in the occurrence and development of ESCC. However, the details of its function and molecular mechanism are unclear and need to be further explored.
As with many tumors, the occurrence and development of ESCC is related to many signaling pathways, including the Wnt/β-catenin pathway. Both Chinese and foreign studies have confirmed that miR-1 down-regulates the proliferation and migration of breast cancer stem cells by inhibiting the Wnt/β-catenin signaling pathway (9). Recently, miR-98 was reported to inhibit the development of retinoblastoma through mediating the Wnt/β-catenin signaling pathway by targeting HMGA2 (10). In nasopharyngeal carcinoma, miR-506 inhibits tumor growth and metastasis via inhibition of the Wnt/β-catenin signaling pathway through down-regulating LHX2 (11). These miRNAs function biologically in tumor diseases; therefore, we speculated that miR-212-3p may also affect the proliferation and other biological functions of ESCC via the Wnt/β-catenin signaling pathway. This study aimed to investigate this hypothesis to provide new ideas for the treatment of ESCC. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/jtd-20-2558).
Methods
Collection and processing of clinical specimens
Cancer tissue samples and adjacent tissue samples were collected from 60 patients with ESCC who were treated in The First Affiliated Hospital of Nanchang University between 2018 and 2019. The postoperative pathological stage of all patients was determined by three pathologists. The tumor tissues were separated into two parts: one part was preserved with RNA preservation solution, and the other was washed with cold phosphate buffer solution (PBS), treated with diethyl pyrocarbonate (DEPC) to remove the blood, and frozen with liquid nitrogen. None of the patients received preoperative chemotherapy or radiotherapy. For each patient, ESCC was the primary lesion and was confirmed by pathological examination. None of the patients had any history of major systemic disease. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the Medical Ethics Committee of The First Affiliated Hospital of Nanchang University. Signed informed consent was obtained from each patient.
Cell culture
RPMI-1640 medium and 0.25% trypsin were purchased from Gibco, USA. Fetal bovine serum (FBS) was obtained from Hyclone, USA. A total RNA extraction kit was purchased from Tiangen Biotech (Beijing) Co., Ltd. A PrimeScript™ RT regent kit and an SYBR Premix Ex Taq™quantitative real-time PCR (qRT-PCR) kit were purchased from Takara Bio Inc., Japan. Reverse transcription and real-time PCR primers were designed and synthesized by Shanghai GenePharma Co., Ltd. Normal human esophageal epithelial cell line HET-1A and ESCC lines EC9706, KYSE-140, KYSE-150, and TE-10 were acquired from the cell bank of Shanghai Institute of Cell Biology, Chinese Academy of Sciences. The cells were cultured with RPMI-1640 culture solution containing 10% FBS, 100 U/mL penicillin, and 100 µg/mL streptomycin and incubated at 37 °C with 5% CO2 and saturated humidity. The cells were digested with 0.25% trypsin every 2–3 days for passage, and cells in the logarithmic phase were used for subsequent experiments.
Cell transfection
MiR-212-3p mimic, NC mimic, siRNA NC (si-NC), SOX4 siRNA (si-SOX4), and miR-202-3p mimic+overexpressing-SOX4(oe-SOX4) were purchased from RiboBio (Guangzhou, China) and used in strict accordance with the reagent instructions supplied by the company. Transfection was carried out with Lipofectamine 2000. EC9706 cells were divided into five groups: the miR-212-3p mimic group, the NC mimic group, the si-NC group, the si-SOX4 group, and the miR-212-3p + oe-SOX4 group. The EC9706 cells were seeded in 6-well plates at a density of 5×1052 cells/well. When the cell fusion density reached 60–70%, 2 µL of Lipofectamine 2000 and 5 nM miR-212-3p mimic, NC, si-NC, si-SOX4, and miR-212-3p mimic + oe-SOX4 were mixed at a volume ratio of 1:50 and added to the culture medium for transfection. After 6 hours of transfection, the cells were washed three times with PBS, the culture medium was replaced with common culture medium, and then culture continued for a further 48 hours. The transfection efficiency was detected by qRT-PCR.
MTT assay for cell proliferation detection
The cells in the logarithmic growth phase were collected and treated with 0.25% trypsin. After being counted, the cells were cultured in a 96-well culture plate with 5,000 cells per well. Four parallel wells and correction wells without cells were established for each group. The plate was placed in an incubator containing 5% CO2 and 100% humidity at 37 °C for 24 hours for cell adhesion. Then, 20 µL of 5 g/L MTT solution was added to each well, and after incubation for a further 4 hours the supernatant was discarded. Then, 20 µL of DMSO was added, and the solution was shaken gently for 10 min to completely dissolve the crystal substance. The absorption value was measured at 490 nm using an enzyme-linked fluorescence analyzer. The experiment was repeated three times.
Transwell for cell invasion detection
These five groups were established as described above. Complete medium containing 10% FBS was added to the lower chamber of the Transwell. A polycarbonate membrane (13 mm in diameter with 8 µm pore size) was paved between the upper and lower chambers, and 50 µg/well of artificial Matrigel (BD Company) was evenly paved onto the membrane and the Transwell was polymerized in an incubator at 37 °C for 30 min. A total of 400 µL (2×104 cells) of cell suspension in the logarithmic growth phase was added to the upper chamber and cultured for 48 hours at 37 °C, 5% CO2. The fluid was removed from the upper chamber fluid, and the uninvaded cells and Matrigel on the membrane surface were wiped off. The cells were washed in normal saline, fixed in methanol for 30 min, and stained with 0.1% crystal violet. Three parallel samples were set in each group. The number of penetrating cells in each group was randomly counted under a 400× microscopic field, and the mean values were taken. The experiment was repeated three times.
Flow cytometry for apoptosis detection
The treated cells were digested and collected, and 50,000–100,000 cells were centrifuged (1,000 r/min, 5 min). The supernatant was discarded, then 195 µL of Annexin V-FITC binding solution was added and the cells were gently resuspended. Subsequently, 5 µL of Annexin V-FITC reagent was added and mixed gently, and placed at room temperature for 10 min avoiding light. Another 10 µL of propidium iodide (PI) staining solution was added, gently mixed, and placed at room temperature for 10 min avoiding light. Then, the cells were resuspended by gently adding 200 µL of Annexin V-FITC binding solution. The cell suspension were subsequently detected by flow cytometry: green fluorescence was shown after Annexin V-FITC binding and red fluorescence was shown after PI binding.
Luciferase assay
The cells in the logarithmic growth phase were made into cell suspension and inoculated in a 24-well plate. Upon reaching 60% confluency, the cells were transfected according to the following groups using Lipofectamine 2000: Group A: miR-212-3p mimics and wild-type (WT) plasmid containing SOX4 3'-UTR end; Group B: NC mimics and WT plasmid containing SOX4 3'-UTR end; Group C: miR-212-3p mimics and mutant (MUT) plasmid containing SOX4 3'-UTR end; and Group D: NC mimics and MUT plasmid containing SOX4 3'-UTR end. After 48 hours of co-transfection, a dual-luciferase reporter assay system was employed to validate the relative luciferase activity among the different groups. The following calculation formula was used: luciferase activity = firefly luciferase activity/Renilla luciferase activity. The experiment was repeated three times.
Quantitative RT-PCR
Total RNA was isolated from ESCC cell lines (EC9706, KYSE-140, KYSE-150, and TE-10), a normal esophageal epithelial cell line (HET-1A), and tumor tissues using TRIzol reagent (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA). A total of 500 ng RNA was reverse transcribed into cDNA using a cDNA transcription kit (ABI). Transcription was subsequently carried out at 16 °C for 30 min, followed by incubation at 42 °C for 30 min and enzyme inactivation at 85 °C for 5 min. Rapid quantitative PCR was performed using SYBRH Select Master Mix (Invitrogen). Transcription was performed using the following parameters: 16 °C 30 min, 42 °C 30 min, 84 °C 30 min. Quantitative RT-PCR: 95 °C 2 min, 95 °C 15 s, 60 °C 40 cycles. All results were standardized to the expression of glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The 2−ΔΔCt method was used for quantitative analysis. The primer sequences used were as follows: miR-212-3p, 5'-AGCATCCACGAGCAAGAGAC-3' and 5'-GATGCTACTAGTGTGGCGGG-3'; U6, 5'-GCTTCGGCAGCACATATACTAAAAT-3' and 5'-CGCTTCACGAATTTGCGTGTCAT-3'; SOX4, 5'-CTTGACATGATTAGCTGGCATGATT-3' and 5'-CCTGTGCAATATGCCGTGTAGA-3'; GAPDH, 5'-AACGGATTTGGTCGTATTG-3' and 5'-GGAAGATGGTGATGGGATT-3'.
Western blot to detect the expression level of SOX4 and related proteins
At 48 hours after transfection, the cells were lysed and total protein was extracted. After the protein content had been determined, the protein was isolated using SDS-PAGE, then transferred to a polyvinylidene difluoride (PVDF) membrane, blocked with Tris Buffered saline Tween (TBST) buffer containing 5% skim milk powder. After washing, the membrane was incubated with the following primary antibodies at a dilution of 1:1,000 overnight at 4 °C: rabbit monoclonal antibody SOX4 (ab243041), rabbit polyclonal antibody Wnt1(ab15251), rabbit monoclonal antibody β-catenin (ab32572), rabbit monoclonal antibody c-Myc (ab32072), rabbit monoclonal antibody Cyclin D1 (ab134175), and rabbit monoclonal antibody GAPDH (ab181602). Following that, the membrane was incubated with horseradish peroxidase (HRP)-labeled secondary antibody for 2 hours. The color was developed by chemiluminescence, and the results were analyzed using a gel imaging system. Images were subsequently quantified with Image J (National Institutes of Health).
Statistical analysis
All data obtained from the experiments were statistically analyzed using SPSS 22.0 software (International Business Machines Corporation, Armonk, New York, USA). The measurement data were expressed as mean ± standard deviation (
Results
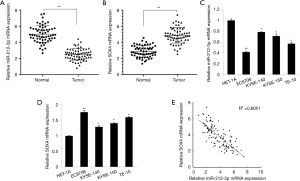
Negative correlation between miR-212-3p and SOX4 expression in ESCC
The quantitative polymerase chain reaction (qPCR) assay results showed that the expression of miR-212-3p was significantly decreased and that of SOX4 was significantly increased in ESCC tissues compared with adjacent tissues (Figure 1A,B). This finding suggested that miR-212-3p and SOX4 were involved in the development of ESCC. To further verify this discovery, with human normal esophageal epithelial cells (HET-1A group) used as a control group, qRT-PCR revealed that miR-212-3p was down-regulated, while SOX4 was up-regulated, in the ESCC cell lines EC9706, KYSE-140, and TE-10 (Figure 1C,D). Correlation analysis revealed that the expression levels of miR-212-3p and HOXA5 were significantly negatively correlated (Figure 1E). The expression of miR-212-3p was lowest in EC9706 cells and the expression of SOX4 was highest in EC9706 cells. Therefore, the EC9706 cell line was selected for subsequent experiments.
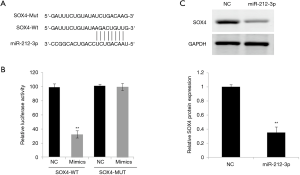
miR-212-3p targeted regulation SOX4 expression
First, a significant binding site between miR-212-3p and SOX4 was predicted using the bioinformatics software Starbase V2.0 (Qu Lab. School of Life Science, Sun Yat-sen University, China) (Figure 2A). To further verify the target regulation relationship between miR-212-3p and SOX4, WT or MUT plasmid containing SOX4 3'-UTR end and miR-212-3p mimics or NC were co-transfected into EC9706 cells to detect luciferase activity in Group A, B, C and D, respectively. As shown in Figure 2B, following co-transfection, luciferase activity was significantly inhibited. The results of western blot showed that the protein expression level of SOX4 inEC9706 cells was significantly reduced when miR-212-3p was overexpressed compared with the NC group (Figure 2C). This finding suggested that miR-212-3p could regulate the level of SOX4 expression by binding to SOX4 3'-UTR.
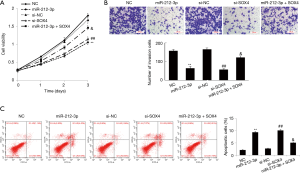
miR-212-3p regulates SOX4 expression to inhibit the proliferation and invasion and to promote apoptosis of ESCC cells
To investigate the biological function of miR-212-3p in ESCC cells, gain- and loss-of-function experiments were performed. EC9706 cells were transfected with miR-212-3p mimic, NC MIMIC, si-NC and si-SOX4 and miR-212-3p mimic + oe-SOX4. After successful transfection, proliferation, invasion, and apoptosis of EC9706 cells were detected by MTT assay, Transwell assay, and flow cytometry, respectively. The results showed that compared with NC group, the cell proliferation rate and cell invasion ability were significantly decreased in the miR-212-3p mimics group, while apoptosis was significantly increased (P<0.05). Further, compared with the si-NC group, the cell proliferation rate and cell invasion ability were significantly decreased in the si-SOX4 group, while apoptosis was significantly increased (P<0.05). Compared with miR-212-3p group, the cell proliferation rate and cell invasion ability were significantly increased in the miR-212-3p + oe-SOX4 group, while apoptosis was significantly decreased (P<0.05). No significant difference was observed between the NC group and the si-NC group (P>0.05) (Figure 3).
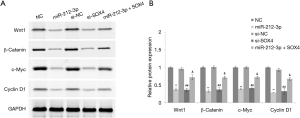
Effect of miR-212-3p targeting SOX4 on the Wnt/β-catenin signaling pathway in EC9706 cells
In order to verify miR-212-3p inhibits the Wnt/β-catenin signaling pathway to affect proliferation, invasion, apoptosis, and other biological functions in ESCC, cells were transfected with miR-212-3p mimic, NC mimic, si-NC, si-SOX4, and miR-212-3p mimic + oe-SOX4.After transfection, the expression levels of the corresponding proteins of the Wnt/β-catenin signaling pathway(Wnt1, β-catenin, c-Myc, and Cyclin D1) were detected. The results of western blot showed that the protein levels of Wnt1, β-catenin, c-Myc, and Cyclin D1 in the miR-212-3p group were significantly decreased compared with those in the NC group (P<0.05). Further, the protein levels of Wnt1, β-catenin, c-Myc, and Cyclin D1 in the si-SOX4 group were significantly decreased compared with those in the si-NC group (P<0.05). Meanwhile, the protein levels of Wnt1, β-catenin, c-Myc, and Cyclin D1 in the miR-212-3p + oe-SOX4 group were significantly increased compared with those in the miR-212-3p group (P<0.05). No significant difference was observed between the NC group and the si-NC group (P>0.05) (Figure 4). These results suggested that miR-212-mediated the proliferation, invasion, apoptosis, and other biological functions of ESCC through inhibiting the Wnt/β-catenin signaling pathway by targeting SOX4.
Discussion
With the continuous improvements in molecular biology and oncology, our understanding of the pathogenesis of ESCC has developed; however, the exact etiology and pathogenesis of the disease are not entirely clear (12). ESCC is known to be a multi-factor and multi-stage complex biological process. Its occurrence and development are associated with the abnormal expression and functions of related genes. It is essential that the exact mechanisms of ESCC are explored in depth, so that early biomarkers for its diagnosis, treatment, and prognosis can be identified and targeted drugs can be designed, thus improving the clinical diagnosis and treatment for patients with the disease (13). Despite the discovery and confirmation of a large number of ESCC-related genes, more thorough investigations of the mechanisms underlying signal pathway disorder and abnormal gene expression regulation in ESCC need to be carried out (14). In recent years, miRNAs have been found to be closely related to, and thus play an important role in, the occurrence and development of ESCC through their regulation of corresponding signaling pathways (15). MiRNAs are a class of short non-coding RNAs (18–25 nucleotides in length), which bind to the 3'-UTR of target mRNAs to act as post-transcriptional regulators of gene expression (16). Therefore, finding ESCC-related miRNAs and conducting in-depth studies on their functions and regulatory mechanisms is of great importance.
A significant amount of research on the relationship between miRNA and ESCC has been conducted. In various studies in which ESCC tissues and esophageal tissues were paired, or peripheral blood samples from ESCC patients and healthy controls were paired, an abnormal expression of specific miRNA was found in ESCC. The expression of miR-21, a typical tumor-associated miRNA, is elevated in various tumor tissues, including lung, breast, and cervical cancer (17). Xie et al. found that the miR-21 expression level in the salivary supernatant of ESCC patients was significantly increased compared with healthy controls, and that its sensitivity and specificity for diagnosing ESCC were 84.4% and 62.5%, respectively (18,19). Zhang et al. also discovered that miR-21 expression was significantly increased in cancer tissues and peripheral serum from ESCC patients. The area under the receiver operating curve of miR-21 serum was 0.847, which indicates that it has a degree of value for the diagnosis of ESCC (20). Other miRNAs reported to be up-regulated in ESCC tissues are miR-25, miR-208, miR-223, and miR-296. On the contrary, some miRNAs such as miR-143, miR-195, miR429. miR-138, and miR-628, are down-regulated in ESCC tissues and serum, and may have inhibitory effect on ESCC (21-24).
MiR-212-3p was recently discovered to be associated with tumor proliferation and metastasis (25). Overexpression of mir-212-3p is associated with the prognosis of ESCC (26). However, its function in ESCC is unclear. The results of this study revealed that miR-212-3p expression was significantly decreased in human ESCC tissues and ESCC cell lines, indicating that miR-212-3p might be involved in the development and progression of ESCC. Further comparative analysis of databases found that miR-212-3p had binding sites in the 3'-UTR of SOX4, and there was a significant negative correlation between the two, which suggests that miR-212-3p might be involved in the occurrence and development of ESCC through its regulation of SOX4 expression. There are 32 members in the SOX gene family, of which the role of SOX4 in tumors is of particular interest. The SOX gene family controls development and plays an important role in sex differentiation and the formation of tissue and organ during ontogeny. In meta-analysis, SOX4 is one of 64 “tumor markers” in human, and plays an important role in tumor occurrence and progression (27). Studies have found that SOX4 is highly expressed in liver cancer tissue samples (28), and down-regulation of SOX4 can significantly inhibit the growth and metastasis of liver cancer (29). In recent years, a considerable number of studies have reported the overexpression of SOX4 in various tumors, such as prostate (30), breast (31), and lung cancer (32). The overexpression of SOX4 can promote cell survival and transformation by promoting proliferation and inhibiting apoptosis, suggesting that SOX4 is an oncogene in tumor development. Moreover, SOX4 has also been confirmed to be closely related to tumor development and metastasis. Recent studies have pointed out that the expression of SOX4 in gastric cancer, lung cancer, and endometrial cancer is closely related to tumor differentiation and metastasis (33). For example, in hepatocellular carcinoma, overexpression of SOX4 can not only promote the invasion and migration of cancer cells, but also inhibit the pro apoptotic effect mediated by p53 by combining with p53 promoter, thus reducing the effect of radiotherapy (34). In bladder cancer, overexpression of SOX4 can promote the apoptosis of cancer cells, thus play a role in tumor inhibition (35). Progesterone can induce SOX4 expression in breast cancer (36). In hepatoma cells, PGA2 and m-pgj2 can promote the expression of SOX4 (37). SOX4 was up-regulated in SOX4 and SOX4 in endometrial cells. In prostate cancer, SOX4 protein can bind to its own promoter and regulate its expression (38). This indicates that SOX4 overexpression may be one of the important factors in tumor development.
In this study, we found that the overexpression of SOX4 protein in ESCC tissues was closely related to the metastasis of ESCC, while the expression of miR-212-3p was significantly down-regulated in ESCC tissues. Luciferase activity analysis also confirmed that the SOX4 gene was a direct target of miR-212-3p. In this study, exogenous miR-212-3p mimic was transfected into EC9706 cells and the changes in SOX4 protein levels after up-regulation of miR-212-3p were observed in vitro and in vivo. The SOX4 protein expression levels in cells transfected with miR-212-3p mimic were significantly down-regulated compared with the control group. This result supported the findings of previous experimental studies and confirmed the regulatory effect of miR-212-3p on SOX4 protein expression. To further investigate the regulatory effect of miR-212-3p on biological functions in ESCC by targeting SOX4, we performed gain- and loss-of-function experiments and found that overexpression of miR-212-3p and interference with SOX4 significantly inhibited the proliferation and invasion of EC9706 cells, and promoted apoptosis. Overexpression of SOX4 reversed the functional effects of up-regulation of miR-212-3p in EC9706 cells. To some extent, miR-212-3p was demonstrated to target and regulate SOX4 expression to affect the occurrence and development of ESCC.
After determining the effect of miR-212-3p on the biological characteristics of ESCC by targeting SOX4, our next focus was to elucidate the specific mechanism of this effect. A large number of studies have confirmed that the Wnt/β-catenin signaling pathway plays a key role in various cell events, including the regulation of gene expression, growth, and proliferation. Cell proliferation and metastasis have been found to be vital in the occurrence and development of cancer. The Wnt/β-catenin and P13K/AKT pathways are important signaling pathways in the regulation of cell proliferation and metastasis. These pathways are often abnormally activated in the development of many cancers, including prostate cancer, oral squamous cell carcinoma, and cutaneous squamous cell carcinoma (39-41). Wnt1, β-catenin, c-Myc, and Cyclin D1 are the key factors in the Wnt/β-catenin signaling pathway (42). We found that up-regulation of miR-212-3p expression and down-regulation of SOX4 expression inhibited the protein expression of Wnt1, β-catenin, c-Myc, and Cyclin D1 (P<0.05). Furthermore, after up-regulation of miR-212-3p, overexpression of SOX4 inhibited the protein expression of Wnt1, β-catenin, c-Myc, and Cyclin D1. Therefore, we speculated that miR-212-3p inhibits the Wnt/β-catenin signaling pathway to mediate the biological functions of ESCC by targeting SOX4.
In conclusion, miR-212-3p expression is low in ESCC cells and tissues. After in-depth investigation of the mechanism, it was confirmed that miR-212-3p regulated the biological functions such as proliferation, invasion and apoptosis of ESCC cells through SOX4. However, there are still some deficiencies in this study, such as insufficient clinical sample size and lack of follow-up animal experiment verification. We will try our best to study and confirm the reliability of the molecular mechanism in the follow-up study. At present, there are a large number of studies related to the vivo study of miRNAs in ESCC. This study suggests that miR-212-3p may be a potential cancer target, and also demonstrates the clinical potential of miR-212-3p as a target for prognosis for patients with ESCC.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/jtd-20-2558
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jtd-20-2558
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-20-2558). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013), and was approved by the Medical Ethics Committee of The First Affiliated Hospital of Nanchang University. Signed informed consent was obtained from each patient.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- de Castro G Junior, Segalla JG, de Azevedo SJ, et al. A randomised phase II study of chemoradiotherapy with or without nimotuzumab in locally advanced oesophageal cancer: NICE trial. Eur J Cancer 2018;88:21-30. [Crossref] [PubMed]
- Zhao C, Yang J, Xu L. The hOGG1 Ser326Cys polymorphism and esophageal cancer risk: a meta-analysis of 1,875 cancer cases and 3,041 controls. Ann Transl Med 2019;7:438. [Crossref] [PubMed]
- Zhao H, Diao C, Wang X, et al. LncRNA BDNF-AS inhibits proliferation, migration, invasion and EMT in oesophageal cancer cells by targeting miR-214. J Cell Mol Med 2018;22:3729-39. [Crossref] [PubMed]
- Xie SH, Lagergren J. Risk factors for oesophageal cancer. Best Pract Res Clin Gastroenterol 2018;36-37:3-8. [Crossref] [PubMed]
- Chai T, Shen Z, Chen S, et al. Right versus left thoracic approach oesophagectomy for oesophageal cancer: a systematic review and meta-analysis protocol. BMJ Open 2019;9:e030157. [Crossref] [PubMed]
- Ren LH, Chen WX, Li S, et al. MicroRNA-183 promotes proliferation and invasion in oesophageal squamous cell carcinoma by targeting programmed cell death 4. Br J Cancer 2014;111:2003-13. [Crossref] [PubMed]
- Ide S, Toiyama Y, Shimura T, et al. MicroRNA-503 promotes tumor progression and acts as a novel biomarker for prognosis in oesophageal cancer. Anticancer Res 2015;35:1447-51. [PubMed]
- Chen Z, Liu Y, Qi B, et al. MicroRNA-212 facilitates the motility and invasiveness of esophageal squamous carcinoma cells. Mol Med Rep 2019;20:3633-41. [PubMed]
- Liu T, Hu K, Zhao Z, et al. MicroRNA-1 down-regulates proliferation and migration of breast cancer stem cells by inhibiting the Wnt/β-catenin pathway. Oncotarget 2015;6:41638-49. [Crossref] [PubMed]
- Li W, Wang J, Zhang D, et al. MicroRNA-98 targets HMGA2 to inhibit the development of retinoblastoma through mediating Wnt/β-catenin pathway. Cancer Biomark 2019;25:79-88. [Crossref] [PubMed]
- Liang TS, Zheng YJ, Wang J, et al. MicroRNA-506 inhibits tumor growth and metastasis in nasopharyngeal carcinoma through the inactivation of the Wnt/β-catenin signaling pathway by down-regulating LHX2. J Exp Clin Cancer Res 2019;38:97. [Crossref] [PubMed]
- Shen F, Chen J, Guo S, et al. Genetic variants in miR-196a2 and miR-499 are associated with susceptibility to esophageal squamous cell carcinoma in Chinese Han population. Tumour Biol 2016;37:4777-84. [Crossref] [PubMed]
- Tu Y, Tan F, Zhou J, et al. Pristimerin targeting NF-κB pathway inhibits proliferation, migration, and invasion in esophageal squamous cell carcinoma cells. Cell Biochem Funct 2018;36:228-40. [Crossref] [PubMed]
- Kim R, Keam B, Kwon D, et al. Programmed death ligand-1 expression and its prognostic role in esophageal squamous cell carcinoma. World J Gastroenterol 2016;22:8389-97. [Crossref] [PubMed]
- Song X, You W, Zhu J, et al. A Genetic Variant in miRNA-219-1 Is Associated with Risk of Esophageal Squamous Cell Carcinoma in Chinese Kazakhs. Dis Markers 2015;2015:541531.
- Wu C, Wang C, Guan X, et al. Diagnostic and prognostic implications of a serum miRNA panel in oesophageal squamous cell carcinoma. PLoS One 2014;9:e92292. [Crossref] [PubMed]
- Xia Y, Wang Y, Wang Q, et al. Increased miR-203-3p and reduced miR-21-5p synergistically inhibit proliferation, migration, and invasion in esophageal cancer cells. Anticancer Drugs 2019;30:38-45. [Crossref] [PubMed]
- Winther M, Alsner J, Tramm T, et al. Evaluation of miR-21 and miR-375 as prognostic biomarkers in esophageal cancer. Acta Oncol 2015;54:1582-91. [Crossref] [PubMed]
- Xie ZJ, Chen G, Zhang XC, et al. Saliva supernatant miR-21: a novel potential biomarker for esophageal cancer detection. Asian Pac J Cancer Prev 2012;13:6145-9. [Crossref] [PubMed]
- Zhang T, Zhao D, Wang Q, et al. MicroRNA-1322 regulates ECRG2 allele specifically and acts as a potential biomarker in patients with esophageal squamous cell carcinoma. Mol Carcinog 2013;52:581-90. [Crossref] [PubMed]
- Li S, Li Z, Guo F, et al. miR-223 regulates migration and invasion by targeting Artemin in human esophageal carcinoma. J Biomed Sci 2011;18:24. [Crossref] [PubMed]
- Wang TY, Zhang QQ, Zhang X, et al. The effect of recombinant lentiviral vector encoding miR-145 on human esophageal cancer cells. Tumour Biol 2015;36:9733-8. [Crossref] [PubMed]
- Wu C, Li M, Hu C, et al. Clinical significance of serum miR-223, miR-25 and miR-375 in patients with esophageal squamous cell carcinoma. Mol Biol Rep 2014;41:1257-66. [Crossref] [PubMed]
- Wang Y, Li M, Zang W, et al. MiR-429 up-regulation induces apoptosis and suppresses invasion by targeting Bcl-2 and SP-1 in esophageal carcinoma. Cell Oncol (Dordr) 2013;36:385-94. [Crossref] [PubMed]
- Zhang L, Huang LS, Chen G, et al. Potential Targets and Clinical Value of MiR-224-5p in Cancers of the Digestive Tract. Cell Physiol Biochem 2017;44:682-700. [Crossref] [PubMed]
- Qi B, Liu SG, Qin XG, et al. Overregulation of microRNA-212 in the poor prognosis of esophageal cancer patients. Genet Mol Res 2014;13:7800-7. [Crossref] [PubMed]
- Rhodes DR, Yu J, Shanker K, et al. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc Natl Acad Sci U S A 2004;101:9309-14. [Crossref] [PubMed]
- Hur W, Rhim H, Jung CK, et al. SOX4 overexpression regulates the p53-mediated apoptosis in hepatocellular carcinoma: clinical implication and functional analysis in vitro. Carcinogenesis 2010;31:1298-307. [Crossref] [PubMed]
- Liao YL, Sun YM, Chau GY, et al. Identification of SOX4 target genes using phylogenetic footprinting-based prediction from expression microarrays suggests that overexpression of SOX4 potentiates metastasis in hepatocellular carcinoma. Oncogene 2008;27:5578-89. [Crossref] [PubMed]
- Yang M, Wang J, Wang L, et al. Estrogen induces androgen-repressed SOX4 expression to promote progression of prostate cancer cells. Prostate 2015;75:1363-75. [Crossref] [PubMed]
- Song GD, Sun Y, Shen H, et al. SOX4 overexpression is a novel biomarker of malignant status and poor prognosis in breast cancer patients. Tumour Biol 2015;36:4167-73. [Crossref] [PubMed]
- Wang D, Hao T, Pan Y, et al. Increased expression of SOX4 is a biomarker for malignant status and poor prognosis in patients with non-small cell lung cancer. Mol Cell Biochem 2015;402:75-82. [Crossref] [PubMed]
- Shen R, Pan S, Qi S, et al. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 in gastric cancer. Biochem Biophys Res Commun 2010;394:1047-52. [Crossref] [PubMed]
- Aaboe M, Birkenkamp-Demtroder K, Wiuf C, et al. SOX4 expression in bladder carcinoma: clinical aspects and in vitro functional characterization. Cancer Res 2006;66:3434-42. [Crossref] [PubMed]
- Zhang J, Liang Q, Lei Y, et al. SOX4 induces epithelial-mesenchymal transition and contributes to breast cancer progression. Cancer Res 2012;72:4597-608. [Crossref] [PubMed]
- Milne GL, Zanoni G, Porta A, et al. The cyclopentenone product of lipid peroxidation, 15-A2t-isoprostane, is efficiently metabolized by HepG2 cells via conjugation with glutathione. Chem Res Toxicol 2004;17:17-25. [Crossref] [PubMed]
- Saegusa M, Hashimura M, Kuwata T. Sox4 functions as a positive regulator of β-catenin signaling through upregulation of TCF4 during morular differentiation of endometrial carcinomas. Lab Invest 2012;92:511-21. [Crossref] [PubMed]
- Jafarnejad SM, Ardekani GS, Ghaffari M, et al. Pleiotropic function of SRY-related HMG box transcription factor 4 in regulation of tumorigenesis. Cell Mol Life Sci 2013;70:2677-96. [Crossref] [PubMed]
- Pak S, Park S, Kim Y, et al. The small molecule WNT/β-catenin inhibitor CWP232291 blocks the growth of castration-resistant prostate cancer by activating the endoplasmic reticulum stress pathway. J Exp Clin Cancer Res 2019;38:342. [Crossref] [PubMed]
- Lee SH, Koo BS, Kim JM, et al. Wnt/β-catenin signalling maintains self-renewal and tumourigenicity of head and neck squamous cell carcinoma stem-like cells by activating Oct4. J Pathol 2014;234:99-107. [Crossref] [PubMed]
- Wu N, Du Z, Zhu Y, et al. The Expression and Prognostic Impact of the PI3K/AKT/mTOR Signaling Pathway in Advanced Esophageal Squamous Cell Carcinoma. Technol Cancer Res Treat 2018;17:1533033818758772. [Crossref] [PubMed]
- Yoshida T, Sopko NA, Kates M, et al. Three-dimensional organoid culture reveals involvement of Wnt/β-catenin pathway in proliferation of bladder cancer cells. Oncotarget 2018;9:11060-70. [Crossref] [PubMed]
(English Language Editor: J. Reynolds)


