
An artificial neural network model predicting pathologic nodal metastases in clinical stage I–II esophageal squamous cell carcinoma patients
Introduction
Esophageal cancer (EC) is one of the most common malignant tumours in humans with poor prognosis. The overall 5-year survival rate of patients with EC ranges from 15% to 25% (1). According to the Global Cancer Statistics of 2018, among the 36 cancer diseases in 185 countries surveyed, the incidence of EC ranked seventh among all cancer diseases, and the mortality rate ranked sixth (2). Two common histologic subtypes have marked geographic distributions: squamous cell carcinoma (SCC) and adenocarcinoma (AC). In China, more than 90% of patients with EC are eventually diagnosed with SCC (3).
Treatment strategies for esophageal squamous cell carcinoma (ESCC) include surgery, radiotherapy, chemotherapy and etc. Different treatment strategies depend on the tumour pathologic stages. Precision medicine requires us to classify different states and processes of a disease accurately to achieve personalized treatment strategies for patients. However, due to the inaccuracy of clinical staging, significant percentages of patients were under-staged or over staged (4-6). Under-staged patients who should receive preoperative neoadjuvant chemoradiotherapy underwent surgery directly (7); over-staged patients received unnecessary induction therapy but did not improve long-term outcomes (8). Preoperative clinical staging is essential for developing rational treatment strategies that determine whether patients should receive preoperative neoadjuvant therapy or not. Therefore, in order to obtain the most suitable treatment strategies, it is vital to improve the accuracy of clinical staging.
The new eighth edition of the American Joint Committee on Cancer (AJCC) Cancer Staging system separates the clinical staging system from the pathologic staging system. The new clinical staging system is grouped by histopathologic cell type; the two main cell types of EC (SCC and AC) have independent staging standards. However, current clinical staging based largely on imaging and biomarker information is limited by the resolution and accuracy of each technique and remains coarse and imprecise (mainly when assessing clinical N staging) (9-12). The aim of this study was to construct and validate an artificial neural network (ANN) model to improve the accuracy of clinical N staging in patients with ESCC. We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/jtd-20-1956).
Methods
Study design and settings
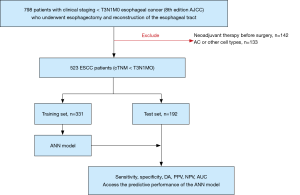
This is a retrospective observational study design. A total of 798 patients with clinical staging I–II (cTNM < T3N1M0) EC (eighth edition AJCC) who underwent esophagectomy and reconstruction of the esophageal tract in Jinling Hospital of Nanjing Medical University from January 2017 to April 2019 were retrospectively enrolled in this study. Exclusion criteria included: (I) patients who received neoadjuvant therapy before surgery (n=142); (II) patients who had AC or other cell types (n=133). The remaining 523 cases consisted of 415 men and 108 women.
Collection of data
Preoperative examination for clinical staging included oesophagogastroscopy and biopsy to determine location, histologic subtypes, and grade; endoscopic ultrasound (EUS) and chest and abdominal computed tomography (CT) for cT; EUS fine-needle aspiration (EUS-FNA) for cN (non-peritumoral); positron emission tomography-CT (PET-CT) for cM and cN (9). Clinical N stage was established using a combination of EUS-FNA and PET-CT. Criteria for N+: (I) EUS-FNA positive cases were defined as those whose cytological diagnosis confirmed the presence of malignant cells in the aspirated lymph node consistent with origin from ESCC. All EUS-FNAs were performed with a CookTM 22-gauge needle by a single experienced endosonographer. (II) The criterion of positive lymph nodal metastasis for PET was determined to be more than 2.5 the maximum standardized uptake values (SUVmax) (type of scanner: Philips Gemini TF 16, Philips, The Netherlands; amount of tracer agent: 3.7 MBq; slice thickness of CT: 3 mm/slice). SUVmax for each site was calculated by the conventional formula normalized for body weight. All PET/CT images were evaluated by two experienced nuclear medicine physicians, and the diagnoses were made by consensus.
The pathologic staging was confirmed by pathologic examination of the tumour tissue and lymph node resected during surgery in all patients. All of the patients agreed to participate and provided informed consent. This study was reviewed and approved by the Institutional Review Board of the Jinling Hospital (approval number 2015NZKY-028-03). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). A flowchart depicting the process used during the study is shown in Figure 1.
ANN modelling and validation
According to the principle of simple random sampling, the 523 cases were divided into two groups (training set, 350 patients; test set, 173 patients). The training set was used to construct the ANN prediction models, and the test set was used to verify and evaluate the accuracy of the ANN models.
A total of 28 variables including the baseline characteristics, perioperative data, and pathologic features of the 523 ESCC patients were collected and summarised in this study. Univariable analysis was performed to assess the candidate variables associated with pathologic lymph node metastasis among the 28 variables; then the candidate variables were input into the input layer to construct the ANN prediction models. The standard feed-forward, back-propagation neural network (BPNN) consisted of three layers composed of logical units: an input layer, a hidden layer, and an output layer. Beginning with an input layer consisting of all of the patients in the training set, the ANN models, similar to the hierarchical synaptic organisation of neurons in the brain, assigned each candidate variable an importance (weight) (13). The hidden layer received the weighted synapses and synthesised the data with the specified activation function; the output layer accepted the hidden layer and calculated the results. In this study, Sigmoid and Softmax were used as the activation functions of the hidden layer and output layer, respectively. The number of neurons in the hidden layer of the ultimate prediction model depended on the predictive performance and clinical relevance of the ANN model.
The diagnostic performance of the ANN model was assessed using the test set. To evaluate the accuracy of the prediction model, we compared the predicted outcomes of the ANN model with the actual observations and then calculated the overall sensitivity (SEN), specificity (SPE), false positive and negative rates, positive predictive value (PPV), negative predictive value (NPV), and diagnostic accuracy (DA) of the model. Receiver operating characteristic (ROC) curves were generated as primary measures of each model’s discriminant ability.
Statistical analysis
We compared the classification and continuous variables between the training set and the test set to ensure that the two groups were statistically comparable. The categorical variables were analysed using the χ2 or Fisher’s exact test, as appropriate. The Shapiro-Wilk test was used to assess whether the continuous data were normally distributed. On the basis of the results of the Shapiro-Wilk test, the continuous variables were compared using Student’s t-test or Mann-Whitney non-parametric tests, as appropriate. Univariable analysis and ANN modelling were performed using SPSS version 21.0 (IBM Corp., Armonk, NY, USA). P values <0.05 were defined as statistically significant.
Results
Study population and characteristics
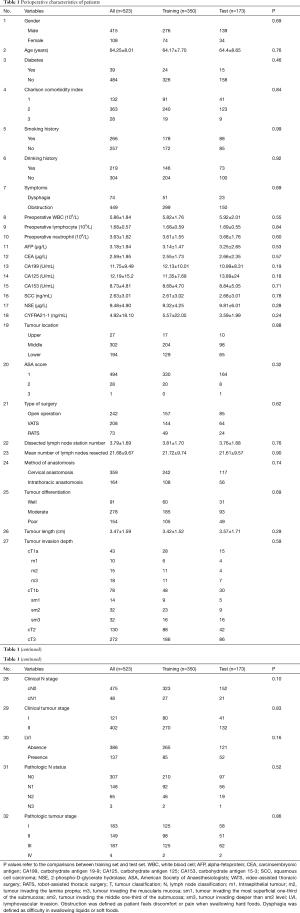
A total of 523 patients suspected to have limited local-regional ESCC (clinical staging I–II) participated in this study. The pathologic nodal status distribution was as follows: N0: 307 (58.7%), N1: 148 (28.3%), N2: 65 (12.4%) and N3: 3 (0.5%). The 41.3% of patients were confirmed pathologic nodal positive (216/523). The demographics of all patients are illustrated in Table 1. According to the results of the univariate analysis, there were no statistical differences between the two sets in the 32 variables enrolled (P>0.05, Table 1), indicating that training set and test set were statistically comparable. Mean dissected lymph node station number in 523 patients were 3.79±1.69; mean number of lymph nodes resected were 21.68±9.67. There is no statistical difference in SUVmax between clinically unrecognized positive lymph nodes and pathologically negative lymph nodes (1.73±0.66 vs. 1.68±0.59, P=0.62).
Full table
ANN modelling in the training set
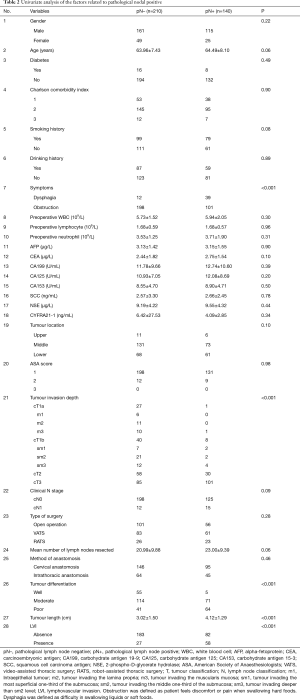
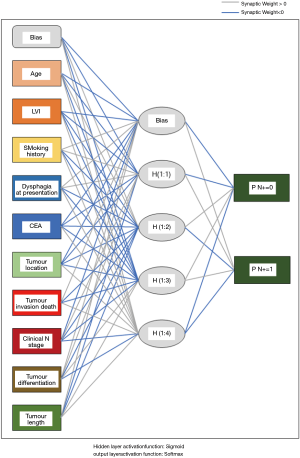
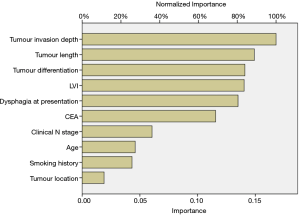
As illustrated in Table 2, the univariate analysis was performed among 28 variables to identify the candidate variables using the data from the training set. Of the 28 variables, a total of 10 variables were identified as candidate variables [including age, smoking history, dysphagia, carcinoembryonic antigen (CEA), tumour location, tumour invasion depth, clinical N stage, tumour differentiation, tumour length and lymphovascular invasion (LVI), P<0.20]. The 10 variables were input into the input layer as neurons to construct the ANN prediction model. The eventual ANN prediction model is shown in Figure 2. When the hidden layer was set to four, the ANN model obtained a good prediction performance. According to the ANN model generated by SPSS, we obtained the normalised importance of each variable (Figure 3). Tumour invasion depth, tumour length, tumour differentiation, LVI and dysphagia were identified as important predictors for predicting pathologic nodal positive patients among all 10 variables (normalised importance >80%); their normalised importance values were 100%, 89.3%, 83.5%, 83.0% and 80.2%, respectively.
Full table
Validation of the ANN model
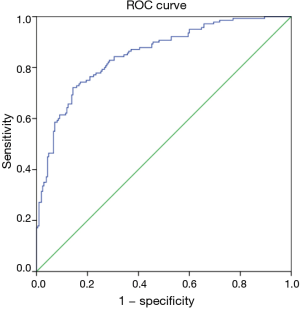
We verified the performance of the ANN model in the training and test sets, respectively. The results are summarised in Table 3. The ROC curve is illustrated in Figure 4. The area under the ROC curve (AUC) value for the ANN model verified in the test set was 0.852.

Full table
Discussions
Over the past few decades, despite the continuous updating of the AJCC staging system, the clinical staging of EC patients still lacks sufficient accuracy. Several studies have reported the inaccuracy of the clinical staging. Zhang et al. reported that more than 50% of cT2N0 patients were under-staged based on the 6th edition of the AJCC staging system (7). Worrell et al. evaluated the accuracy of the seventh edition of the AJCC staging system; of 88 AC patients enrolled with clinical stage ≤ T3N1M0, 21 patients were diagnosed with final pathologic stage > T3N1M0 (14). In 2018, a systematic review and meta-analysis reported similar results. Lv et al. collected eight retrospective studies of 2,646 patients with cT2N0M0 (neoadjuvant therapy combined with surgery group, n=961; simple surgery group, n=1,685). The results showed that tumour staging was under-estimated in six studies; further subgroup analysis suggests that N staging was more likely to be under-estimated (30.1% vs. 35.1%) than T staging (6). This was because the current preoperative examination is inaccurate (especially for N staging) and equipment has regional differences. To date, previous studies have found several predictors related with pN+. Gaur et al. found that cT (OR, 5.6; 95% CI, 1.7 to 18.6; P<0.01) and tumour length (OR, 7.0; 95% CI, 2.7 to 18.1; P<0.001) were significant independent factors of nodal metastases in patients with esophageal AC (EAC) (15). Barbetta enrolled 80 cT2N0M0 EAC patients and found vascular invasion was an independent risk factor for nodal disease (16). Samson et al. enrolled 46 ESCC patients (29.3%) and 111 EAC patients (70.7%); LVI (OR, 6.0; 95% CI, 2.9 to 12.5; P<0.001) and tumour grade 3 (OR, 9.4; 95% CI, 1.8 to 48.4; P=0.007) were identified as independent predictors for nodal metastases (11). Currently, data on patients with ESCC are lacking. In this study, we constructed an ANN model for predicting pathologic nodal positive ESCC patients. Based on the subsequent validation, the ANN model was proven to have a good performance and may help clinicians improve the clinical strategies.
The ANN is a statistical model that mimics the structure and function of biological neural networks (17). Compared with traditional statistical techniques (for example, logistic regression and Cox proportional hazards regression), the self-learning ability of the BPNN (by modifying weights and thresholds based on new inputs) makes it possible to approximate any non-linear function with arbitrary precision (18,19). This machine learning statistical method is increasingly used in the medical field, such as for disease diagnosis and the prediction of patient outcomes or complications (20-22). This is also the first study demonstrating the application of BP ANNs for predicting pN+ patients with ESCC.
In this study, 523 patients with ESCC (clinical TNM classification < T3N1M0) were enrolled and randomly assigned to a training group (n=350) and a test group (n=173). 9% (48/523) of the patients were diagnosed with clinical nodal positive preoperatively; whereas 41.3% (216/523) of the patients were confirmed pathologic nodal positive. This result may imply the inaccuracy of the current clinical examination for N staging. Ideally, clinical staging should correctly determine a patient’s lymph node metastasis. To achieve this goal, an ANN model was established based on the baseline characteristics, perioperative data, and pathologic features in the training set (Figure 2). As illustrated in Figure 3, tumour invasion depth, tumour length, tumour differentiation, LVI and dysphagia were identified as important factors influencing model decision (the normalised importance values were 100%, 89.3%, 83.5%, 83.0% and 80.2%, respectively). Subsequently, we performed a series of verifications on the constructed prediction model based on the test set. The AUC revealed that the ANN model had good predictive accuracy (SEN, 82.5%; SPE, 78.9%; PPV, 77.9%; NPV, 83.3%; DA, 80.9%; AUC, 0.852).
The eighth edition of the AJCC staging system separates clinical staging from pathologic staging and establishes independent clinical staging strategies for AC and ESCC, respectively (9). The eighth edition of the AJCC cancer clinical staging system is based solely on T staging and N staging and does not take into account tumour differentiation, tumour length, LVI, or symptoms. In this study, the above four indicators were identified as important predictors influencing model decision. We suggest that future clinical staging criterion can consider incorporating the above indicators to make the subgroups more homogeneous.
Several drawbacks limited our study. Because only patients with clinical staging < T3N1M0 were enrolled (patients with advanced stages were excluded), both SEN and accuracy in the detection of nodal metastasis may have been relatively low. The ANN model constructed in this study can be used as a supplement to the current clinical staging system to reduce the false negative rate of preoperative lymph node assessment (if the patient is diagnosed as clinical stage I–II, we can use the model to screen out patients with suspicious N+). This single-centre study is also limited by its retrospective nature and the small number of patients. In addition, the ANN model constructed in this study also lacks external validation. In the future, we will continue to expand the sample size (using clinical databases or collecting multi-centre clinical data) to improve the versatility and accuracy of the model.
Conclusions
In conclusion, tumour invasion depth, tumour length, tumour differentiation, LVI and dysphagia were important factors for predicting pathologic nodal positive. The ANN model integrates these factors and appears suitable for clinical staging stratification for ESCC. The prediction model is based on easily available perioperative indicators and can have potential implications for clinical management.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/jtd-20-1956
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jtd-20-1956
Peer Review File: Available at http://dx.doi.org/10.21037/jtd-20-1956
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-20-1956). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was reviewed and approved by the Institutional Review Board of the Jinling Hospital (approval number 2015NZKY-028-03). All of the patients agreed to participate and provided informed consent. This study is based on data retrieved from a hospital medical record system. All personal data have been protected and secured according to current national and international laws.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Pennathur A, Gibson MK, Jobe BA, et al. Oesophageal carcinoma. Lancet 2013;381:400-12. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Speicher PJ, Ganapathi AM, Englum BR, et al. Induction therapy does not improve survival for clinical stage T2N0 esophageal cancer. J Thorac Oncol 2014;9:1195-201. [Crossref] [PubMed]
- Chen WH, Chao YK, Chang HK, et al. Long-term outcomes following neoadjuvant chemoradiotherapy in patients with clinical T2N0 esophageal squamous cell carcinoma. Dis Esophagus 2012;25:250-5. [Crossref] [PubMed]
- Lv HW, Xing WQ, Shen SN, et al. Induction therapy for clinical stage T2N0M0 esophageal cancer: A systematic review and meta-analysis. Medicine (Baltimore) 2018;97:e12651. [Crossref] [PubMed]
- Zhang JQ, Hooker CM, Brock MV, et al. Neoadjuvant chemoradiation therapy is beneficial for clinical stage T2 N0 esophageal cancer patients due to inaccurate preoperative staging. Ann Thorac Surg 2012;93:429-35; discussion 436-7. [Crossref] [PubMed]
- Dijksterhuis WPM, Hulshoff JB, van Dullemen HM, et al. Reliability of clinical nodal status regarding response to neoadjuvant chemoradiotherapy compared with surgery alone and prognosis in esophageal cancer patients. Acta Oncol 2019;58:1640-7. [Crossref] [PubMed]
- Rice TW, Gress DM, Patil DT, et al. Cancer of the esophagus and esophagogastric junction-Major changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J Clin 2017;67:304-17.
- Jiang C, Chen Y, Zhu Y, et al. Systematic review and meta-analysis of the accuracy of 18F-FDG PET/CT for detection of regional lymph node metastasis in esophageal squamous cell carcinoma. J Thorac Dis 2018;10:6066-76. [Crossref] [PubMed]
- Samson P, Puri V, Robinson C, et al. Clinical T2N0 esophageal cancer: identifying pretreatment characteristics associated with pathologic upstaging and the potential role for induction therapy. Ann Thorac Surg 2016;101:2102-11. [Crossref] [PubMed]
- Shin S, Kim HK, Choi YS, et al. Clinical stage T1-T2N0M0 oesophageal cancer: accuracy of clinical staging and predictive factors for lymph node metastasis. Eur J Cardiothorac Surg 2014;46:274-9; discussion 279. [Crossref] [PubMed]
- Bertolaccini L, Solli P, Pardolesi A, et al. An overview of the use of artificial neural networks in lung cancer research. J Thorac Dis 2017;9:924-31. [Crossref] [PubMed]
- Worrell SG, Alicuben ET, Oh DS, et al. Accuracy of clinical staging and outcome with primary resection for local-regionally limited esophageal adenocarcinoma. Ann Surg 2018;267:484-8. [Crossref] [PubMed]
- Gaur P, Sepesi B, Hofstetter WL, et al. A clinical nomogram predicting pathologic lymph node involvement in esophageal cancer patients. Ann Surg 2010;252:611-7. [PubMed]
- Barbetta A, Schlottmann F, Nobel T, et al. Predictors of nodal metastases for clinical T2N0 esophageal adenocarcinoma. Ann Thorac Surg 2018;106:172-7. [Crossref] [PubMed]
- Chatzimichail E, Matthaios D, Bouros D, et al. gamma -H2AX: a novel prognostic marker in a prognosis prediction model of patients with early operable non-small cell lung cancer. Int J Genomics 2014;2014:160236. [Crossref] [PubMed]
- Chen HL, Yu SJ, Xu Y, et al. Artificial neural network: a method for prediction of surgery-related pressure injury in cardiovascular surgical patients. J Wound Ostomy Continence Nurs 2018;45:26-30. [PubMed]
- Li H, Luo M, Zheng J, et al. An artificial neural network prediction model of congenital heart disease based on risk factors: A hospital-based case-control study. Medicine (Baltimore) 2017;96:e6090. [Crossref] [PubMed]
- Zhang X, Yang Y, Wang Y, et al. Detection of the BRAF V600E mutation in colorectal cancer by NIR spectroscopy in conjunction with counter propagation artificial neural network. Molecules 2019;24:2238. [Crossref] [PubMed]
- Zhao Y, Zhao L, Mao T, et al. Assessment of risk based on variant pathways and establishment of an artificial neural network model of thyroid cancer. BMC Med Genet 2019;20:92. [Crossref] [PubMed]
- Jiang N, Xu X. Exploring the survival prognosis of lung adenocarcinoma based on the cancer genome atlas database using artificial neural network. Medicine (Baltimore) 2019;98:e15642. [Crossref] [PubMed]


