
Impact of EGFR amplification on survival of patients with EGFR exon 20 insertion-positive non-small cell lung cancer
Introduction
Lung cancer is one of the most common malignant diseases worldwide and a major cause of cancer-related death (1,2). Around 85% of all lung cancers are non-small cell lung cancers (NSCLCs), among which adenocarcinoma and squamous cell carcinoma (SCC) are the most common types (3,4). During the past decade, the development of new technologies such as next-generation sequencing (NGS) has enhanced our understanding of how the genetic characteristics of NSCLC influence its pathogenesis and drug-resistance (5). The use of modern molecular biology tools to study the genetic changes underlying NSCLC has allowed us to embark on an era of precision medicine, and as a result the one-size-fits-all therapeutic model has been abandoned. It is now known that mutations in the gene encoding the epidermal growth factor receptor (EGFR) promote tumor proliferation and metastasis, and the use of EGFR-tyrosine kinase inhibitors (EGFR-TKIs) as a targeted therapy has markedly improved the management of patients with NSCLC (5-7).
In general EGFR-TKIs are more effective for NSCLC with activating mutations of EGFR than for NSCLC with wild-type EGFR (8). Among the known EGFR gene mutations, L858R and exon 19 deletion (EGFR 19del) account for the majority (85–90%) of mutations in the EGFR kinase domain that respond well to EGFR-TKIs (9). However, not all EGFR mutations are sensitive to EGFR-TKIs. The insertion of three or more nucleotides into exon 20 of EGFR (EGFR ex20ins) is a known driver for NSCLC. Most EGFR ex20ins mutations are resistant to the currently available EGFR-TKIs and associated with a poor prognosis (10,11). For example, it has been reported that the response rates of patients with EGFR ex20ins to first-generation (erlotinib and gefitinib) and second-generation (afatinib) EGFR-TKIs are low, ranging from 0–8.7% (12,13). Although new drugs targeting NSCLC with EGFR ex20ins mutations, such as poziotinib, TAK-788 and TAS6417, have been developed, these agents are still being evaluated in clinical trials (14-16). Thus, effective treatment options for EGFR ex20ins mutations are limited.
EGFR ex20ins is usually mutually exclusive from certain other disease-driving gene mutations (such as EGFR 19del, EGFR L858R, and mutations in ERBB2, ALK, BRAF and RET) but is often accompanied by mutations in TP53, EGFR amplification, CDKN2A and CDKN2B (17). Among the many known EGFR ex20ins mutation subtypes, patients with EGFR p.A763_Y764insFQEA respond to EGFR-TKIs (18), and this has driven research into identifying other EGFR ex20ins mutation subtypes that are sensitive to EGFR-TKIs. However, the reality is that clinical outcomes will likely differ between two patients with the same EGFR ex20ins mutation and the same treatment protocol, in part because of the influence of other accompanying gene mutations. Although EGFR amplification is present in 22% of patients with EGFR ex20ins (17), no previous studies have evaluated the effect of EGFR amplification on the clinical outcomes of patients with EGFR ex20ins mutations treated with EGFR-TKIs or/and chemotherapy.
The aim of this study was to investigate whether EGFR amplification affects the progression-free survival (PFS) and overall survival (OS) of patients with EGFR ex20ins mutations treated with an EGFR-TKI and/or other chemotherapeutic agent. We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/jtd-20-1630).
Methods
Study design and participants
This is an observational longitudinal cohort study. This prospective case series included consecutive treatment-naïve adult patients with advanced NSCLC who were seen at Guangdong Provincial People’s Hospital between November 2017 and February 2019. The participants were enrolled from a prospectively managed database that also collected information regarding the patients’ clinical characteristics, NGS data, treatment and clinical outcomes. The inclusion criteria were: (I) age ≥18 years; (II) diagnosis of advanced NSCLC; (III) no previous treatment for NSCLC; and (IV) EGFR ex20ins mutation subsequently confirmed by NGS of peripheral blood and/or tissue specimens. Patients with incomplete or missing data were excluded from the final analysis. The study investigators played no role in the design or implementation of the treatment protocols. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics and Scientific Committees of Guangdong Provincial People’s Hospital (GDREC2016175H) and informed consent was taken from all the patients.
Grouping
The distribution of the EGFR ex20ins mutation subtypes was analyzed for all patients. Patients were divided into an EGFR amplification group and a non-EGFR amplification group. Since the presence/absence of EGFR amplification could only be determined by NGS of tumor tissue, the clinical characteristic and the survival analysis (PFS and OS) only included patients who provided tumor tissue samples. Furthermore, patients were excluded from the survival analysis if they had EGFR mutations known to be sensitive to EGFR-TKIs (19del, L858R, G719X, L861Q and S768I). For the survival analysis, TKI subgroup including patients with once or more TKI treatment and Chemotherapy subgroup including patients with once or more chemotherapy treatment.
Genomic analysis
NGS was performed by Burning Rock Biotech (Guangzhou, China), which is a Clinical Laboratory Improvement Amendments-certified testing center. The samples for NGS included plasma and tumor tissue. For the extraction of cell-free DNA (cfDNA) from plasma, 10 mL of whole blood was collected in K3EDTA-containing tubes (Cell-Free DNA BCT; Streck, La Vista, NE, USA) and centrifuged at 2,000 g for 10 min at 4 °C within 72 h of collection. The supernatant was transferred to a 15-mL centrifuge tube and centrifuged at 16,000 g for 10 min at 4 °C, and the supernatant was transferred to a new tube for storage at −80 °C until further use. Circulating cfDNA was extracted using the QIAamp Circulating Nucleic Acid kit (Qiagen, Hilden, Germany) and quantified using the dsDNA HS Assay Kit and Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA). For plasma genotyping, NGS of peripheral white blood cells was also performed to avoid false positive plasma genotyping due to clonal hematopoiesis. The extraction of DNA from tissue samples was achieved using the QIAamp DNA FFPE Tissue Kit (Qiagen) in accordance with the manufacturer’s instructions. The DNA concentration was determined using the Qubit dsDNA Assay (Life Technologies).
Samples were analyzed using panels of 31, 61, 295 or 520 cancer-related genes. Burrows-Wheeler Aligner 0.7.10 was used to map the sequencing data (average sequencing depth, 10,000X). Local alignment optimization, variant calling and annotation (minimum locus depth, 100) were carried out using GATK 3.2, MuTect (Broad Institute, Cambridge, MA, USA) and VarScan (Genome Institute, Washington University, USA). Insertions/deletions and single nucleotide variations (SNVs) were verified using at least two and eight supporting reads, respectively. Single-nucleotide polymorphisms (variants with a population frequency >0.1%) were excluded from further analysis, and the remaining variants were annotated with ANNOVAR and SnpEff v3.6. TopHat2 (Center for Computational Biology, Johns Hopkins University and Genome Sciences Department, University of Washington, USA) and Factera 1.4.3 were utilized for DNA translocation analysis.
Gene copy number variation (CNV) was evaluated with in-house scripts of tumor tissues. Coverage data were corrected for sequencing bias arising from GC content and probe design. The coverage of the various samples was normalized to comparable scales using the average coverage of all captured regions. Gene copy number (CN) at each capture interval was calculated from the ratio of the coverage depth in detected circulating tumor DNA to the average coverage of ≥50 reference samples without CNV. CNV was determined using two criteria: (I) coverage of >60% capture intervals of the genes differed significantly from reference samples (P<0.005 for hotspot genes and P<0.001 for other genes, z-test comparing the coverage of each capture interval to the mean coverage of the interval in all control samples); (II) CN attained the minimal threshold for gain (>2.25 for hotspot genes and >2.5 for others) or loss (<1.75 for hotspot genes and <1.5 for others). CNV distribution plots were utilized to differentiate amplifications from polysomies. EGFR amplification was defined as >2.75 in the 520 panel and >2.25 in the other panels (19).
Collection of clinical data
The following clinical data were extracted from the database: sex, age, smoking history, tumor pathological type, treatment history and the date of occurrence of tumor progression or death.
Outcomes
All patients were followed-up until death or January 8, 2020, whichever occurred first, and the median follow-up time was 630 days. OS was defined as the period from the initial systemic treatment to death of the patient due to any cause. PFS was defined as the period from the initiation treatment to progression of the tumor or death.
Statistical analysis
The data were analyzed using SPSS 22.0 (IBM Corp., Armonk, NY, USA) and Prism 7.0 (GraphPad, San Diego, CA, USA). Quantitative data are expressed as median (range) and were compared between groups using the Mann-Whitney U test. Count data are expressed as n (%) and were compared between groups using Fisher’s exact test. Survival was analyzed using the Kaplan-Meier method and log-rank test. All statistical tests were two-sided, and P<0.05 was considered statistically significant.
Results
Clinical characteristics of the study participants
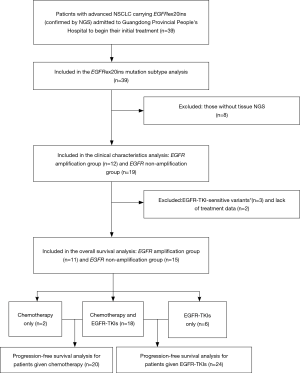
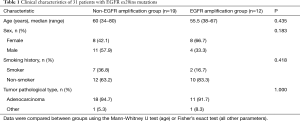
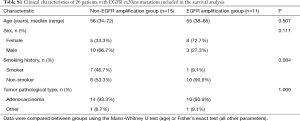
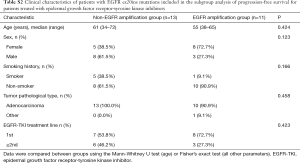
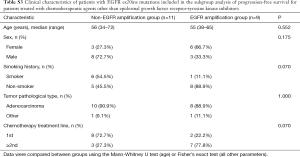
A total of 39 patients were included in the EGFR ex20ins mutation subtypes analysis. For the clinical characteristic analysis, 8 patients were excluded because NGS of tumor tissue samples was not performed. For the survival analysis, 3 patients were excluded because they also had EGFR-TKI-sensitive EGFR mutations (two carrying L858R and one carrying G719X), and 2 patients were excluded due to missing data. Therefore, a total of 26 patients were included in the overall survival analysis (Figure 1). As shown in Table 1, there were no significant differences between the EGFR amplification group (n=12) and non-EGFR amplification group (n=19) in sex, age, smoking history or tumor pathological type. Twenty-six EGFR ex20ins patients included in survival analysis. There were no significant differences between the EGFR amplification group (n=11) and non-EGFR amplification group (n=15) in sex, age, smoking history or tumor pathological type were (Table S1). For the subgroup analyses, 24 of the 26 patients were treated with at least one EGFR-TKI (EGFR amplification group, n=11; non-EGFR amplification group, n=13), and 20 of the 26 patients were administered another type of chemotherapeutic agent (EGFR amplification group, n=9; non-EGFR amplification group, n=11). For each of these subgroup analyses, there were no significant differences between the EGFR amplification group and non-EGFR amplification group in sex, age, smoking history or tumor pathological type (Tables S2 and S3).
Full table
Full table
Full table
Full table
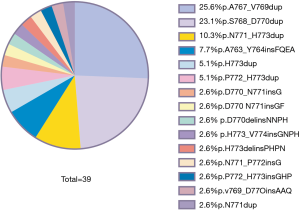
Distribution of EGFR ex20ins mutation subtypes
Fifteen different EGFR ex20ins mutation subtypes were identified in the 39 patients (Figure 2). The three most common EGFR ex20ins subtypes were p.A767_D770dup (25.6%), p.S768_D770dup (23.1%) and p.N771_H773dup (10.3%).
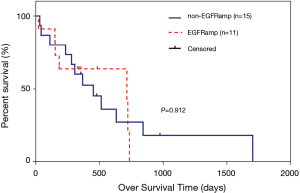
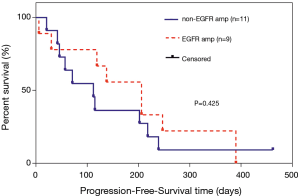
Effects of EGFR amplification on the OS and PFS of patients carrying EGFR ex20ins mutations
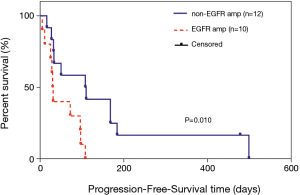
In the overall analysis, there were no significant differences between the EGFR amplification group (n=11) and non-EGFR amplification group (n=15) in median OS (715 vs. 452 days, P=0.912; Figure 3). Among the 20 patients who received at least one chemotherapeutic agent, there were no significant differences between the EGFR amplification group (n=9) and non-EGFR amplification group (n=11) in median PFS (206 vs. 112 days, P=0.425; Figure 4). Among the 24 patients administered at least one EGFR-TKI, the EGFR-TKI was used as a first-line treatment in 8 of the 11 patients (72.7%) in the EGFR amplification group and 7 of the 13 patients (53.8%) in the non-EGFR amplification group (P=0.423, Tables S2). Notably, median PFS was significant longer in the non-EGFR amplification group than in the EGFR amplification group (110 vs. 31 days, P=0.030; Figure 5). Since the EGFRp.A763_Y764insFQEA mutation is sensitive to EGFR-TKIs (18), we repeated the analysis after excluding 2 patients carrying this mutation: median PFS remained significantly longer in the non-EGFR amplification group than in the EGFR amplification group (109 vs. 30.5 days, P=0.010; Figure 6).

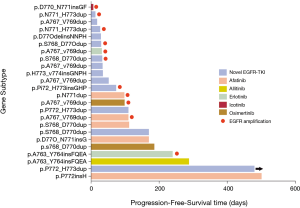
Types of EGFR-TKIs used
Among the 24 patients who received EGFR-TKIs, 5 patients were treated with afatinib, 2 patients were given erlotinib, 2 patients were administered osimertinib, 1 patient was treated with icotinib, 1 patient was given allitinib, and 13 patients were administered a new type of EGFR-TKI as part of a clinical trial (Figure 7). PFS exceeded 1 year in 2 patients: a 66-year-old male with stage IV lung cancer carrying the EGFRp.P772insH mutation who was administered afatinib as a first-line treatment (PFS of 499 days) and a 64-year-old female with stage IV lung cancer carrying the EGFRp.P772_H773dup mutation who was administered a novel EGFR-TKI as a first-line treatment (PFS of 478+ days). Neither of these two patients had co-occurring EGFR amplification.
Discussion
This study demonstrates the EGFR ex20ins gene profile and the impact of EGFR amplification on survival of patients with EGFR ex20ins-positive NSCLC. Patients with NSCLC exhibiting both EGFR ex20ins mutation and EGFR amplification had a significantly shorter PFS than patients with NSCLC exhibiting EGFR ex20ins mutation without EGFR amplification when an EGFR-TKI was used as the treatment but not when another chemotherapeutic drug was used as the treatment. To our knowledge, this is the first report to analyze the association between EGFR amplification and clinical outcome in patients with EGFR ex20ins mutations treated with EGFR-TKIs.
There are many different EGFR ex20ins mutations, and a recent study of 14,483 NSCLC specimens from mainly Caucasian patients identified 64 EGFR ex20ins mutations (17). The three most common mutations detected in the above study were p.A767_D770dup (21%), p.S768_D770dup (20%) and p.N771_H773dup (8%) (17), which is in good agreement with our findings. Although the overall rate of EGFR mutations in NSCLC may be higher in Chinese patients than in Caucasian patients, ethnicity appears to have little effect on the rates of the common EGFR ex20ins mutations.
Previous research has indicated that EGFR ex20ins mutations are generally mutually exclusive from certain other disease-driving genes such as mutations in ERBB2, ALK, BRAF and RET as well as other EGFR mutations (17). However, EGFR ex20ins mutations are usually accompanied by CDKN2A mutations and amplifications of TP53 and EGFR (17). It is worth noting that the rate of EGFR amplification in NSCLC carrying EGFR ex20ins mutations was only 22% in the study by Riess et al. (17) compared with 38.7% in our study. There are three possible explanations for the apparent difference between our findings and those of Riess et al.: (I) the study by Riess et al. included mainly Caucasian patients, who are known to have a lower EGFR mutation rate than Asian patients; (II) the sample size in the study by Riess et al. (n=263) was much larger than that of our study (n=31); and (III) the NGS methods and definition of EGFR amplification differed between the two studies. With regard to the latter point, EGFR amplification was defined as >6 copy number (CN) in the study by Riess et al. and as CN reaching the minimum threshold (>2.75 in the 520 panel and >2.25 in the other panels) in our study; this difference in definition may have contributed to the lower proportion of patients considered to have EGFR amplification in the study by Riess et al. It should be noted that we have previously used fluorescence in situ hybridization (FISH) to validate the NGS methods used to detect EGFR amplification in this study (19). Since there are many NGS testing methods, each with its own definition of EGFR amplification, more research is needed to obtain a consensus threshold for the definition of EGFR amplification.
This study found that PFS was significantly shorter in patients with EGFR amplification than in those without EGFR amplification after their first treatment with an EGFR-TKI, suggesting a less favorable therapeutic effect in patients with EGFR ex20ins mutations who also have EGFR amplification. However, EGFR amplification status did not have a significant effect on PFS in patients given their first chemotherapy. Previous reports have described an association between EGFR amplification and the therapeutic effects of EGFR-TKIs and anti-EGFR monoclonal antibody (20-22). The therapeutic effect of an EGFR-TKI is achieved mainly through inhibition of the EGFR signaling pathway, whereas the benefits of other types of chemotherapy may not be directly related to suppression of EGFR signaling. Therefore, EGFR amplification may only attenuate the therapeutic effects of drugs that block the EGFR signaling pathway.
Anti-EGFR monoclonal antibody can inhibit EGFR signaling, and its combination with cetuximab may be a good treatment option for patients with EGFR ex20ins mutation with EGFR amplification. In a study of 4 patients carrying EGFR ex20ins mutations, the combination of afatinib with cetuximab achieved a partial response in 2 of 3 patients with EGFR CN 1–2 and in another patient with EGFR CN 3–4, and PFS was 2.7, 4.4, 6.4+ (treatment ongoing) and 17.6 months for these patients (23). These results indicate that the combination of an EGFR-TKI with cetuximab may achieve a better therapeutic effect in patients with NSCLC exhibiting both an EGFR ex20ins mutation and EGFR amplification. Future therapeutic strategies for patients with EGFR ex20ins mutations will depend on the results of clinical trials of various new drugs. For now, our first priority is to decide how to optimize the treatment protocol using the drugs currently available on the market.
There are still many limitations in our study. First, the EGFR-TKI treatment regimens varied between patients. The overall efficacy of EGFR-TKIs were limited, and there were obvious differences in sensitivity between individuals. Second, this was a single-center study with a small sample size. Thus, our conclusion was just a suggestive and trending one and larger sample size studies are needed. Patient recruitment is ongoing to validate our preliminary findings.
In summary, there might be a tendency that EGFR amplification is associated with a poorer PFS in patients with EGFR ex20ins-positive NSCLC treated with EGFR-TKIs. Screening for EGFR ex20ins and EGFR amplification with NGS might help to identify patients who would benefit from therapy with an EGFR-TKI. Further large-scale clinical trials with better drugs targeting EGFR ex20ins are needed to confirm our findings.
Acknowledgments
The authors thank the patients, their families and the study personnel who participated in this trial.
Funding: This work was supported by the National Key R&D Program of China (No. 2016YFC1303800 to Q Zhou), the National Natural Science Foundation of China (No. 81871891 to Q Zhou), the High-level Hospital Construction Project (No. DFJH201810 to Q Zhou) and Key Lab System Project of Guangdong Science and Technology Department-Guangdong Provincial Key Lab of Translational Medicine in Lung Cancer (Grant No. 2017B030314120, to YL WU).
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/jtd-20-1630
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jtd-20-1630
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-20-1630). QZ reports speaker fees from AstraZeneca, and Roche. YLW reports speaker fees from AstraZeneca, Eli Lilly, Pfizer, Roche, and Sanofi. WZZ reports speaker fees from AstraZeneca and Roche. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics and Scientific Committees of Guangdong Provincial People’s Hospital (GDREC2016175H) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Barta JA, Powell CA, Wisnivesky JP. Global Epidemiology of Lung Cancer. Ann Glob Health 2019;85:8. [Crossref] [PubMed]
- Molina JR, Yang P, Cassivi SD, et al. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc 2008;83:584-94. [Crossref] [PubMed]
- Beadsmoore CJ, Screaton NJ. Classification, staging and prognosis of lung cancer. Eur J Radiol 2003;45:8-17. [Crossref] [PubMed]
- Loong HH, Kwan SS, Mok TS, et al. Therapeutic strategies in EGFR mutant non-small cell lung cancer. Curr Treat Options Oncol 2018;19:58. [Crossref] [PubMed]
- Yang JC. Osimertinib in pretreated T790M-positive advanced non-small- cell lung cancer: AURA study phase II extension component. J Clin Oncol 2017;35:1288-96. [Crossref] [PubMed]
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. [Crossref] [PubMed]
- Zhao D, Chen X, Qin N, et al. The prognostic role of EGFR-TKIs for patients with advanced non-small cell lung cancer. Sci Rep 2017;7:40374. [Crossref] [PubMed]
- Gazdar AF. Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene 2009;28:S24-S31. [Crossref] [PubMed]
- Arcila ME, Nafa K, Chaft JE, et al. EGFR exon 20 insertion mutations in lung adenocarcinomas: prevalence, molecular heterogeneity, and clinicopathologic characteristics. Mol. Cancer Ther 2013;12:220-9. [Crossref] [PubMed]
- Kobayashi Y, Mitsudomi T. Not all epidermal growth factor receptor mutations in lung cancer are created equal: Perspectives for individualized treatment strategy. Cancer Sci 2016;107:1179-86. [Crossref] [PubMed]
- Yang JC, Sequist LV, Geater SL, et al. Clinical activity of afatinib in patients with advanced non-small-cell lung cancer harbouring uncommon EGFR mutations: a combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol 2015;16:830-8. [Crossref] [PubMed]
- Tu HY, Ke EE, Yang JJ, et al. A comprehensive review of uncommon EGFR mutations in patients with non-small cell lung cancer. Lung Cancer 2017;114:96-102. [Crossref] [PubMed]
- Robichaux JP, Elamin YY, Tan Z, et al. Mechanisms and clinical activity of an EGFR and HER2 exon 20–selective kinase inhibitor in non–small cell lung cancer. Nat Med 2018;24:638-46. [Crossref] [PubMed]
- Doebele RC, Riely GJ, Spira AI, et al. First report of safety, PK, and preliminary antitumor activity of the oral EGFR/HER2 exon 20 inhibitor TAK-788 (AP32788) in non–small cell lung cancer (NSCLC). J Clin Oncol 2018;36:9015. [Crossref]
- Hasako S, Terasaka M, Abe N, et al. TAS6417, a novel EGFR inhibitor targeting exon 20 insertion mutations. Mol Cancer Ther 2018;17:1648-58. [Crossref] [PubMed]
- Riess JW, Gandara DR, Frampton GM, et al. Diverse EGFR exon 20 insertions and co-occurring molecular alterations identified by comprehensive genomic profiling of non-small cell lung cancer. J Thorac Oncol 2018;13:1560-8. [Crossref] [PubMed]
- Lin YT, Liu YN, Wu SG, et al. Epidermal growth factor receptor tyrosine kinase inhibitor-sensitive exon 19 insertion and exon 20 insertion in patients with advanced non-small-cell lung cancer. Clin Lung Cancer 2017;18:324-332.e1. [Crossref] [PubMed]
- Zhang YC, Chen ZH, Zhang XC, et al. Analysis of resistance mechanisms to abivertinib, a third-generation EGFR tyrosine kinase inhibitor, in patients with EGFR T790M-positive non-small cell lung cancer from a phase I trial. EBioMedicine 2019;43:180-7. [Crossref] [PubMed]
- Paz-Ares L, Socinski MA, Shahidi J, et al. Correlation of EGFR-expression with safety and efficacy outcomes in SQUIRE: a randomized, multicenter, open-label, phase III study of gemcitabine–cisplatin plus necitumumab versus gemcitabine–cisplatin alone in the first-line treatment of patients with stage IV squamous non-small-cell lung cancer. Ann Oncol 2016;27:1573-9. [Crossref] [PubMed]
- Wang C, Xu F, Shen J, et al. Successful treatment of lung adenocarcinoma with gefitinib based on EGFR gene amplification. J Thorac Dis 2018;10:E779-E783. [Crossref] [PubMed]
- Hirsch FR, Varella-Garcia M, McCoy J, et al. Increased epidermal growth factor receptor gene copy number detected by fluorescence in situ hybridization associates with increased sensitivity to gefitinib in patients with bronchioloalveolar carcinoma subtypes: a Southwest Oncology Group Study. J Clin Oncol 2005;23:6838-45. [Crossref] [PubMed]
- van Veggel B, de Langen AJ, Hashemi SMS, et al. Afatinib and cetuximab in four patients with EGFR Exon 20 insertion–positive advanced NSCLC. J Thorac Oncol 2018;13:1222-6. [Crossref] [PubMed]


