
The role of angiogenetic single-nucleotide polymorphisms in thymic malignancies and thymic benign lesions
Introduction
Thymic epithelial tumors (TETs) are rare malignancies arising in the anterior superior mediastinum with a global annual incidence of 0.15/100,000 cases (1,2). They represent a heterogeneous group of tumors with an extremely wide spectrum of morphologic features. They show different presentations with a variable and unpredictable evolution ranging between an indolent noninvasive and a highly infiltrative and metastasizing behavior (3-5). Moreover, the same thymomas present a large variability of morphological aspects. These tumors are divided in 5 subgroups according to WHO classification (type A, AB, B1, B2, B3) (6). The International Thymic Malignancy Interest group (ITMIG) identified the Masaoka staging system as the most appropriate one that will be used, considering the-modification included by Detterbeck et al. (7).
Furthermore, a large spectrum of benign proliferative morphological lesions including thymic hyperplasia, thymic cysts and thymolipomas may arise in thymus (8).
Despite several studies in molecular biology investigated various genetic aberrations responsible of thymic tumorigenesis, our knowledge in this field remains poor. Angiogenesis has been focused as a fundamental role for the aforementioned mechanism. Vascular endothelial growth factor (VEGF) represents a proangiogenic and autocrine constituent in thymomas and an immunoregulatory factor in normal thymus (9). Thymoma and thymic carcinoma overexpress both VEGF-A and VEGF-receptors, compared to normal thymus. These angiogenesis factors show a correlation with advanced clinical stages, mainly in thymic carcinomas (10). The chromosome 6 (6p21.1) contains the human VEGF-A gene and the chromosome 4 (4q11-q12) carries VEGFR2 or KDR gene. These genes are highly polymorphic in the population, so single nucleotide polymorphisms (SNPs) have been investigated (11-13). The frequency of these SNPs varies in the population. In addition to VEGF-A and KDR, tumor angiogenesis and cancer progression were associated with other several genes, including VEGFR3 (14), platelet-derived growth factor (PDGF) and PDGFRα (15), hypoxia-inducible factor-1α (HIF-1α) (16), and excision repair cross-complementation group 1 (ERCC1) (17). In the last years, the scientific research focused on translational studies of cellular pathways, in order to identify the biology and the genetic and epigenetic aberrations drivers in thymic malignancies. Furthermore, integrating genomic analyses may lead to the identification of new biomolecular targets.
The main aim of our study was to analyze SNPs mainly related to the angiogenesis to assess their role about susceptibility to develop thymic lesions and clinical outcome.
Methods
Study population
The study population includes patients who underwent complete thymic resections for thymic malignancy or thymic benign lesion (including thymic hyperplasia, thymic cysts, thymolipoma and thymic remnant) at our institution (Università Politecnica delle Marche, Italy) between 1993 and 2013.
Other eligibility criteria included performance status according to Eastern Cooperative Oncology Group (ECOG) classification ≤2; normal organ activity; absence of serious comorbidity. Our study has been approved by the regional ethical committee (Comitato Etico Regionale delle Marche, number of approval document: 214439) and it conforms to the provisions of in accordance with the Helsinki Declaration. All patients signed an informed consent to all the diagnostic and therapeutic procedures.
SNP selection, DNA extraction and genotyping
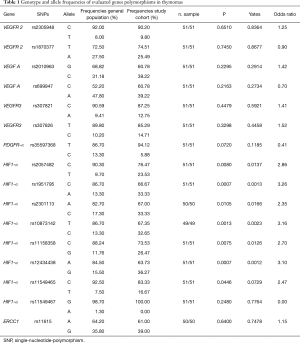
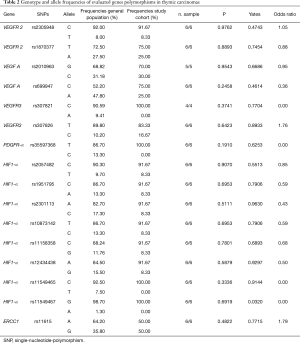
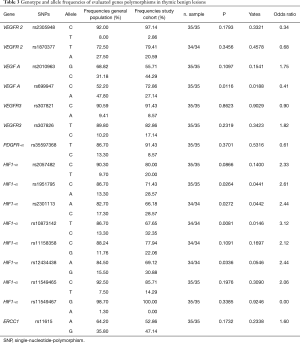
The present study investigated the frequency of SNPs involved in angiogenesis in TETs and in thymic benign lesions also comparing with general population. Tables 1-3 show all frequencies and genotype distributions.
Full table
Full table
Full table
The studied SNPs were carefully chosen through the National Center for Biotechnology Information (NCBI) data, and reviewing available literature, considering the following features:
- The area of the gene; polymorphisms should be located in biologically relevant area (i.e., coding sequence, 5' UTR and 3' UTR or promoter region intron),
- The minor allele frequency (MAF) should be equal to or higher than 10% (with the exception of rs307821, rs11549465 and rs11549767).
Genomic DNA of patient enrolled in the study, was isolated using The RecoverAll™ Total Nucleic Acid Isolation Kit for Formalin Fixed Paraffin-Embedded (FFPE) Tissues (Applied Biosystems, Foster City, CA, USA), according to the manufacturer’s instructions, the DNA extraction has been carried out starting from 30 mg of paraffin-embedded tissue.
To study the genotype of the selected polymorphisms, we used TaqMan SNP Genotyping Assays (Applied Biosystems, Foster City, CA). Samples were loaded on the 7300 Real-Time PCR System (Applied Biosystems) and SDS software v1.4.0 for allelic discrimination (Applied Biosystems), was used to analyzed the amplification product. Some samples were randomly reanalyzed and the results were 100% concordant Frequencies data of CEU population provided by the HapMap project (www.HapMap.org) were taken as reference frequency. If these data were not available, we considered the frequencies reported in the 1000 genome project (www.1000genomes.org).
Statistical analysis
Clinical data were retrospectively collected from medical charts and electronic records. Primary endpoint of the study was to investigate the prognostic role of the selected SNPs.
Overall survival (OS) was defined as the interval between the date of surgery to death or last follow-up visit.
Patients who were not reported as died at the time of the analysis were censored at the date, they were last known to be alive.
Kaplan-Meier method was used to evaluate OS and Mantel-Haenszel log-rank test was applied to compare survival among groups. A Cox-regression model was employed for the univariate analysis and to detect the prognostic role of SNPs. 0.05 value was chosen as statistically significant level.
The Haploview software (Broad Institute, Cambridge, MA) was used to check if the SNPs were in the Hardy–Weinberg equilibrium (HWE) and linkage disequilibrium (LD). In the control subjects, the most common genotypes were considered as references. The chi-square test and a Fisher’s exact probability test was used to assess the association between categorical variables. To adjust the values for multiple comparisons, Benjamin-Hochberg correction method was performed (18).
Statistical analyses were assessed using MedCalc version 11.4.4.0 (MedCalc Software, Broekstraat 52, 9030 Mariakerke, Belgium).
Results
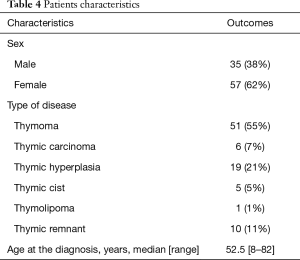
Ninety-two patients were included into the study, M/F ratio was 35/57 and median age was 52.5 years (range, 8–82 years). Clinical characteristics are described in Table 4. Fifty-seven patients presented TETs, including 51 thymomas and 6 thymic carcinomas. In the cohort of patients with TETs, eighteen patients (32%) had Myasthenia Gravis (MG), whilst 3 (5%) presented with lichen ruber planus, pancytopenia, Coombs-positive hemolytic anemia and myositis. Among the 51 patients with thymomas, 29% presented with AB histology, 23% with B2, 16% with A, 16% with B3, 6% with B1, 10% with B2-B3, according to WHO classification. Approximately 63% of the patients had tumors with >5 cm. According to Masaoka-Koga classification, 38% patients presented pathologic stage IIA, while 17%, 31%, 8%, 4%, 2% were found to have stages I, IIB, III, IVA and IVB, respectively.
Full table
All patient underwent total thymectomy for symptomatic mass or MG syndrome.
Description of polymorphisms, HWE and LD
We identified two SNPs in VEGFR2 (rs2305948, rs1870377), VEGF-A (rs2010963, rs699947), and VEGFR3 (rs307821, rs307826), respectively, according to the aforementioned criteria. A single SNP was identified both in PDGFR-α (rs35597368) and in ERCC1 (rs11615) and eight SNPs were identified in HIF1-α (rs2057482, rs1951795, rs2301113, rs10873142, rs11158358, rs12434438, rs11549465, rs11549467). Table 5 summarizes chromosomal location position within the gene, base exchange, main/minor allele frequencies (MAF).
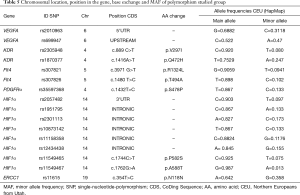
Full table
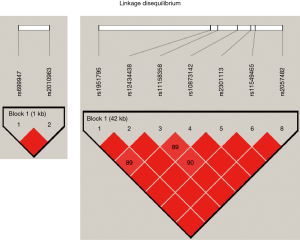
The identified SNPs were in Hardy–Weinberg equilibrium. At the linkage disequilibrium (LD) analysis rs2010963 and rs69947 of VEGF-A were found in strong LD similarity to HIF1-α polymorphisms (Figure 1).
Genotyping and prognostic analyses
Table 1-3 report genotype and allele frequencies of studied SNPs in thymomas, thymic carcinoma and thymic benign lesions respectively.
The SNPs were distributed homogeneously in the entire cohort of patients. Assessing haplotype and allelic frequencies, no statistically significant differences between the prevalence of SNPs in the TETs group and the prevalence of SNPs in the control group were showed. However, we found a significant relationship between the prevalence of SNPs in patients with thymic lesion (thymomas or benign) and in the general population, although no significant association was detected for thymic carcinomas group. This evidence was detected for the following 8 SNPs:
- HIF-1α: rs2057482, rs11549465, rs1951795, rs2301113, rs10873142, rs11158358, rs12434438;
- VEGF-A: rs699947.
In particular, rs2057482, rs11549465 and rs11158358 SNPs of HIF-1α showed significant differences in frequency between the thymomas group and the general population, whereas rs1951795, rs2301113, rs10873142 and rs12434438 SNPs of HIF-1α showed similar differences between both patients’ groups (benign lesion and thymomas) and the general population. Finally, the frequency of rs699947 C polymorphism of VEGF-A showed significant difference between the benign lesions group and general population, whilst no significant difference between patients with malignancy and general population was found.
In particular, the HIF1-α polymorphism rs2057482 C frequency was lower in thymomas (76.5%) in respect of in general population (90.3%), P=0.008, HIF-α allele rs11158358 C was significantly more represented in general population (88.2%) in comparison with patients with thyomas (73.5%), P=0.007; and the frequency of HIF-α allele rs11549465 C was significantly higher in general population (92.5%) respect to thymomas (83.3%), P=0.044.
The frequency of HIF-1α allele rs1951795 C was significantly higher in the general population (86.7%) than study groups, 71.4% in the benign lesions group and 66.7% in thymomas group, P=0.026 and P=0.0007 respectively. Similarly, the frequency of HIF-1α polymorphism rs10873142 T was significantly higher in the general population (86.7%) compared to the study groups (67.7% in benign lesions group; 67.4% in thymomas group; P=0.008 and P=0.001 respectively). Furthermore, HIF-1α allele rs12434438 A was significantly overexpressed in the general population (84.5%) than the study groups (69.1% in benign lesions group; 63.7% in thymomas group; P=0.034 and P=0.0007 respectively). Similarly, the frequency of HIF-1α polymorphism rs2301113 A was significantly higher in the general population (82.7%) than thymomas groups (66.2% in benign lesions group; 67% in thymomas group; P=0.027 and P=0.010 respectively).
Finally, the frequency of VEGF-A polymorphism rs699947 C was significantly higher in patients with benign lesion (72.9%) respect to general population (52.2%), P=0.012, whilst there was no significant difference between patients with malignancy and general population.
Discussion
SNPs analysis is a new promising investigation area in oncology. In fact, SNPs may represent tumor risk markers, prognostic and predictive factors and pharmacological targets.
Angiogenesis plays a crucial role in TETs tumor growth and development, therefore we investigated angiogenesis related genes.
The role of VEGF-A and VEGFR-1 and 2 in thymic proliferative lesions was initially studied by Cimpean and colleagues (9), assessing their immunohistochemical expression in TETs and in other benign conditions. They observed a strong immunostaining for VEGF in type B3 thymomas in neoplastic epithelial cells, in normal endothelial cells, in plasma within the blood vessels and focally in the stroma adjacent to the tumor. VEGFR resulted positive in neoplastic epithelial cells and in endothelium. Thus, they suggested that VEGF may act as an immunoregulatory factor in the normal thymus and as proangiogenic and autocrine factor in thymomas (9,15).
Recently Lattanzio and colleagues (19) conducted a similar study assessing the expression of angiogenesis-related biomarkers in a large series of TETs arranged in Tissue Micro Arrays. The immunostaining of VEGF-A, VEGF-C, VEGF-D, VEGFR-1, VEGFR-2, VEGFR-3, resulted significantly lower in low risk TETs than high risk TETs (thymoma B3 and thymic carcinoma), whilst no differences were observed in terms of PDGFR-β expression.
Several studies showed a correlation between VEGF and VEGFR SNPs and risk of different tumors (20-24).
Nevertheless, the impact of VEGF and VEGFR2-3, HIF-1α, PDGFRα and ERCC1 SNPs on thymic tumor risk and clinical behavior have not been studied, yet. Therefore, the aim of our study was to assess the prognostic role of angiogenesis-related genes SNPs and their susceptibility to develop thymic malignancies. The polymorphism frequencies were analyzed in benign lesions group and malignancy group in comparing each other and with general population.
No statistically significant differences between the prevalence of SNPs in the thymic carcinomas group and the prevalence of SNPs in the general population were showed, probably due to small sample of the study group.
Our results showed that the frequency of HIF-1α polymorphism rs2057482 C, rs11549465 C and rs11158358 C resulted significantly lower in thymomas than in general population. These results suggest the protective role of these SNPs (Figure 2). The role of HIF-1α rs2057482 has not yet been investigated in cancer disease. Conversely, HIF-1α rs11158358 C was considered as risk factor in Chinese women with cervical cancer (25) and rs11549465 C contributes to increased cancer susceptibility according the literature (26).
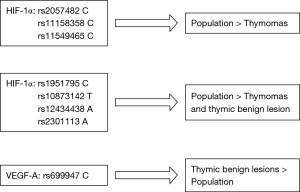
The frequencies of HIF-1α polymorphism rs1951795 C, rs10873142 T, rs1243448 A and rs2301113 A were significantly higher in the general population than in the benign lesions and in the thymomas group, thus suggesting a protective role (Figure 2). This is the first study showing a significant relationship between these alleles and cancer disease. As regard HIF-1α rs11549465 T allele, a correlation with tumor development (27-32).
In our previous study, HIF-1α rs11549465 T frequency was higher in TETs patients vs. general population, although this difference was slightly significant (33).
According to our results, VEGF-A allele rs699947 C seems to be related only to benign lesions suggesting a risk factor role (Figure 2). Several studies considered this allele as risk factor of thyroid cancer and protective factor of colorectal cancer and chronic myeloid leukemia (34).
Within the benign lesion group, we found no significant difference in terms of overall survival between various genotypes. This expected result may depend on the lack of death events in benign lesions groups. In fact, in our previous study on TETs, several polymorphisms resulted significantly associated with better outcome (33).
In the literature, no previous study analyzed the angiogenetic variants in TETs and thymic benign lesion. The data of our research suggest that the angiogenic pathway could have an important role in tumor development according to the first therapeutic experience by Ströbel and colleagues (35), that reported surprising activity of sunitinib metastatic thymic carcinoma patients refractory to conventional treatment.
A phase II study, designed to confirm the activity of Sunitinib in previously treated TETs has also been conducted. This trial by Thomas and colleagues enrolled patients both with thymic carcinoma and thymoma, after least one platinum-based anticancer treatment and with progressive disease. Patients were treated with sunitinib. In 23 evaluable patients with thymic carcinoma and in 16 patients affected by thymoma, the overall response rates (ORR) were 26% and 6%, respectively, and median progression free survivals were 7.2 months for patients with thymic carcinoma and 8.5 months, respectively (36).
Again, Remon and colleagues investigated 28 pretreated TETs patients (20 thymic carcinoma and 8 thymomas) underwent off label Sunitinib. Sixty three percent of disease control rate (86% for thymomas, and 55% for thymic carcinoma) was observed. The median OS was not reached for thymoma patients and for thymic carcinoma patient was instead 12.3 months (P=0.043) (37).
Another Europen phase II trial with the aim of investigation of Sunitinib clinical activity in patients with advanced thymic carcinoma who have failed chemotherapy is ongoing (NCT02623127) (38).
In conclusion, our results suggest possible biomarkers with the SNPs analysis that may be a further asset in definition of patients, who could benefit from anti-VEGF and anti-VEGFR agents thus suggesting the potential importance of innovative antiangiogenic therapies including targeted agents such as Sunitinib, and potentially Sorafenib and Pazopanib. Therefore, this study represents a further step on the road to personalized anticancer therapy for TETs.
Acknowledgments
Funding: This research did not receive any specific grant from any funding agency in the public, commercial or not-for-profit sector. The study was realized with Authors’ University Funding (Università Politecnica delle Marche, Ancona, Italy).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-19-3720). CP serves as an unpaid editorial board member of Journal of Thoracic Disease from Sep 2020 to Aug 2022. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study has been approved by the regional ethical committee (Comitato Etico Regionale delle Marche, number of approval document: 214439). All patients signed an informed consent to all the diagnostic and therapeutic procedures.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Engels EA, Pfeiffer RM. Malignant thymoma in the United States: demographic patterns in incidence and associations with subsequent malignancies. Int J Cancer 2003;105:546-51. [Crossref] [PubMed]
- de Jong WK, Blaauwgeers JL, Schaapveld M, et al. Thymic epithelial tumors: a population-based study of the incidence, diagnostic procedures and therapy. Eur J Cancer 2008;44:123-30. [Crossref] [PubMed]
- Venuta F, Anile M, Diso D. Thymoma and thymic carcinoma. Eur J Cardiothorac Surg 2010;37:13-25. [Crossref] [PubMed]
- Girard N, Mornex F, Van Houtte P, et al. Thymoma: a focus on current therapeutic management. J Thorac Oncol 2009;4:119-26. [Crossref] [PubMed]
- Eng TY, Fuller CD, Jagirdar J, et al. Thymic carcinoma: state of the art review. Int J Radiat Oncol Biol Phys 2004;59:654-64. [Crossref] [PubMed]
- Suster S, Moran CA. Histologic classification of thymoma: the World Health Organization and beyond. Hematol Oncol Clin North Am 2008;22:381-92. [Crossref] [PubMed]
- Detterbeck FC, Nicholson A, Kondo K, et al. The Masaoka-Koga stage classification for thymic malignancies: clarification and definition of terms. J Thorac Oncol 2011;6:S1710-6. [Crossref] [PubMed]
- Rosai J, Levine GD. Atlas of tumor pathology. Tumors of the thymus, Second series, fascicle 13. Armed Forces Institute of Pathology, Washington, DC.
- Cimpean AM, Raica M, Encica S, et al. Immunohistochemical expression of vascular endothelial growth factor A (VEGF), and its receptors (VEGFR1, 2) in normal and pathologic conditions of the human thymus. Ann Anat 2008;190:238-45. [Crossref] [PubMed]
- Lamarca A, Moreno V, Feliu J. Thymoma and thymic carcinoma in the target therapies era. Cancer Treat Rev 2013;39:413-20. [Crossref] [PubMed]
- Vaziri SAJ, Kim J, Ganapathi MK, et al. Vascular endothelial growth factor polymorphisms: role in response and toxicity of tyrosine kinase inhibitors. Curr Oncol Rep 2010;12:102-8. [Crossref] [PubMed]
- Watson CJ, Webb NJ, Bottomley MJ, et al. Identification of polymorphisms within the vascular endothelial growth factor (VEGF) gene: correlation with variation in VEGF protein production. Cytokine 2000;12:1232-5. [Crossref] [PubMed]
- Renner W, Kotschan S, Hoffmann C, et al. A common 936 C/T mutation in the gene for vascular endothelial growth factor is associated with vascular endothelial growth factor plasma levels. J Vasc Res 2000;37:443-8. [Crossref] [PubMed]
- Finnerty H, Kelleher K, Morris GE, et al. Molecular cloning of murine FLT and FLT4. Oncogene 1993;8:2293-8. [PubMed]
- Cimpean AM, Ceauşu R, Encică S, et al. Platelet-derived growth factor and platelet-derived growth factor receptor-α expression in the normal human thymus and thymoma. Int J Exp Pathol 2011;92:340-4. [Crossref] [PubMed]
- Toba H, Kondo K, Sadohara Y, et al. 18F-fluorodeoxyglucose positron emission tomography/computed tomography and the relationship between fluorodeoxyglucose uptake and the expression of hypoxia-inducible factor-1α, glucose transporter-1 and vascular endothelial growth factor in thymic epithelial tumours. Eur J Cardiothorac Surg 2013;44:e105-12. [Crossref] [PubMed]
- Kaira K, Serizawa M, Koh Y, et al. Expression of excision repair cross-complementation group 1, breast cancer susceptibility 1, and β III-tubulin in thymic epithelial tumors. J Thorac Oncol 2011;6:606-13. [Crossref] [PubMed]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J R Stat Soc B Methodological 1995;57:289-300. [Crossref]
- Lattanzio R, La Sorda R, Facciolo F, et al. Thymic epithelial tumors express vascular endothelial growth factors and their receptors as potential targets of antiangiogenic therapy: A tissue micro array-based multicenter study. Lung Cancer 2014;85:191-6. [Crossref] [PubMed]
- Wang K, Liu L, Zhu ZM, et al. Five polymorphisms of vascular endothelial growth factor (VEGF) and risk of breast cancer: a meta-analysis involving 16,703 individuals. Cytokine 2011;56:167-73. [Crossref] [PubMed]
- Chen H, Wang W, Xingjie Z, et al. Association between genetic variations of vascular endothelial growth factor receptor 2 and glioma in the Chinese Han population. J Mol Neurosci 2012;47:448-57. [Crossref] [PubMed]
- Jang MJ, Jeon YJ, Kim JW, et al. Association of VEGF and KDR single nucleotide polymorphisms with colorectal cancer susceptibility in Koreans. Mol Carcinog 2013;52:E60-9. [Crossref] [PubMed]
- Lin L, Cao K, Chen W, et al. Four common vascular endothelial growth factor polymorphisms (−2578C>A, −460C>T, +936C>T, and +405G>C) in susceptibility to lung cancer: a metaanalysis. PLoS One 2013;8:e75123. [Crossref] [PubMed]
- Kämmerer PW, Toyoshima T, Eletr S, et al. Single nucleotide polymorphisms of the vascular endothelial growth factor gene associated with incidence of oral squamous cell carcinoma. J Oral Pathol Med 2010;39:786-92. [Crossref] [PubMed]
- Fu SL, Miao J, Ding B, et al. A polymorphism in the 3' untranslated region of Hypoxia-Inducible Factor-1 alpha confers an increased risk of cervical cancer in a Chinese population. Neoplasma 2014;61:63-9. [Crossref] [PubMed]
- Li HN, He T, Zha YJ, et al. HIF-1α rs11549465 C>T polymorphism contributes to increased cancer susceptibility: Evidence from 49 studies. J Cancer 2019;10:5955-63. [Crossref] [PubMed]
- Tongfeng Z, Jing L, Jangpei Z, et al. Hipoxia-inducible factor1-a gene polymorphism and cancer risk: a meta-analysis. J of Esp & Clin Can Res 2009;28:159. [Crossref]
- Muñoz-Guerra MF. Polymorphism in the hypoxia inducible factor 1 alpha and the impact on the prognosis of early stage of oral cancer. Ann Surg Oncol 2009;16:2351-8. [Crossref] [PubMed]
- Chau CH, Permenter MG, Steinberg SM, et al. Polymorphism in the hypoxia inducible factor 1 alpha gene may confer susceptibility to androgen-independent prostate cancer. Cancer Biol Ther 2005;4:1222-5. [Crossref] [PubMed]
- Orr-Urtreger A, Bar-Shira A, Matzkin H, et al. The homozygouse P582S mutation in the oxygen dependent degradation domain of HIF-1 alpha is associate whit increased risk for prostate cancer. Prostate 2007;67:8-13. [Crossref] [PubMed]
- Lee JY, Choi JY, Lee KM, et al. Rare variant of hypoxia inducible factor 1-alpha (hif-1A) and breast cancer risk in Korean women. Clin Chim Acta 2008;389:167-70. [Crossref] [PubMed]
- Naidu R, Hai YC, Taib NA. Association between hypoxia inducible factor-1alpha (HIF-1A) gene polymorphism and risk of developing breast cancer. Neoplasma 2009;56:441-7. [Crossref] [PubMed]
- Berardi R, Brunelli A, Pagliaretta S, et al. Impact of VEGF, VEGFR, PDGFR, HIF and ERCC1 gene polymorphisms on thymic malignancies outcome after thymectomy. Oncotarget 2015;6:19305-15. [Crossref] [PubMed]
- Lozano-Santos C, Martinez-Velasquez J, Fernandez-Cuevas B, et al. Vascular endothelial growth factor A (VEGFA)gene polymorphisms have an impact on survival in a subgroup of indolent patients with chronic lymphocytic leukemia. PLoS One 2014;9:e101063. [Crossref] [PubMed]
- Ströbel P, Bargou R, Wolff A, et al. Sunitinib in metastatic thymic carcinomas: laboratory findings and initial clinical experience. Br J Cancer 2010;103:196-200. [Crossref] [PubMed]
- Thomas A, Rajan A, Berman AW, et al. Phase II trial of sunitinib in patients with thymic epithelial tumors (TET). J Clin Oncol 2014;32:abstr 7525.
- Remon J, Girard N, Mazieres J, et al. Sunitinib in patients with advanced thymic malignancies: Cohort from the French RYTHMIC network. Lung Cancer 2016;97:99-104. [Crossref] [PubMed]
- Available online: https://www.clinicaltrials.gov/ct2/show/NCT02623127


