
Musashi 2 (MSI2) expression as an independent prognostic biomarker in non-small cell lung cancer (NSCLC)
Introduction
Non-small cell lung cancer (NSCLC) is the leading cause of cancer-related mortality in the world, and metastasis is the most common cause of death in lung cancer patients (1). Despite the recent increase in the number of patients with this disease detected in the earlier stage that can often be treated successfully, they typically have poor prognosis, especially if tumors have begun to disseminate from the original site with a presence of occult metastasis (2). Therefore, it is imperative for the cancer community to develop not only drugs and other treatments effective in advanced stages as well as better adjuvant therapies, but also to search for the novel prognostic and predictive biomarkers in NSCLC.
The evolutionarily conserved translational regulator Musashi-2 (MSI2) is a member of Musashi family of RNA-binding proteins. It regulates mRNA translation of numerous intracellular targets, and influences multiple biological processes, including maintenance of stem cell identity (3). During embryogenesis, MSI2 plays a key role in regulation of asymmetric cell division in Drosophila, and proliferation, and differentiation in the nervous and hematopoietic tissues in mammals (4). MSI2 oncogenic properties were first demonstrated in a murine model of chronic myeloid leukemia in 2010 (5). Musashi proteins overexpression and prognostic significance were demonstrated in several cancer settings, including, but not limited to, colorectal and pancreatic cancers, glioblastomas, and several leukemias (6). Increased expression of MSI2 induced in vitro resistance to paclitaxel in ovarian cancer cells (7). Recently, the role for MSI2 in cancer was redemonstrated in regulation of aggressive breast cancer progression (8) and in supporting transformation of intraepithelial pancreatic neoplasia (PanIN) into ductal pancreatic adenocarcinoma (9).
Previously we reported that MSI2 is present in NSCLC tumors spontaneously arising in transgenic Kras/p53 murine model, upregulated in the metastasis-competent NSCLC murine and human cell lines, overexpressed in primary NSCLC versus normal lung, and progressively elevated in lung cancer patient samples (10). While we observed no correlation with survival in 123 patient NSCLC tissue microarray that mostly consisted of stage I and II patients, we did observe that high levels of MSI2 mRNA are associated with inferior survival (hazard ratio 1.33) in a large NSCLC primary lung cancer samples database (10). In the current study, we performed additional comprehensive correlative analysis between MSI2 expression and clinical follow-up data, using an independent cohort of human NSCLC tissue samples, and different publicly available datasets to determine whether the degree of MSI2 expression varies between clinical-pathological profiles and whether it could be utilized as a novel marker of disease progression. We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/jtd-20-2787).
Methods
Patients
In the current retrospective study, we used randomly selected formalin fixed paraffin embedded (FFPE) NSCLC (n=40) and matching normal tissue specimens (n=20) collected from the Rostov National Medical Research Center Human Tissue Repository Facility (HTRF) in Rostov-on-Don, Russia, and the Republican Oncological Clinical Hospital’s Repository, Kazan, Russia. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the local ethics committees of both organizations. Patients underwent a complete oncological resection and histological verification of tumor at both institutions and provided Institutional Review Board (IRB)—approved informed consents for storing tissue and reviewing of deidentified clinical data at the time of tissue acquisition (IRB No. 8, 02/13/2018 of the local ethics committee of the Kazan Federal University (Kazan, Russia) and IRB No.: 0.1.1.55-16/23/18-8 of National Medical Research Center of Oncology (Rostov-on-Don, Russia). Clinical information (Table 1) from the repository databases was abstracted in an anonymized fashion, and all samples were de-identified.

Full table
Immunohistochemistry of human NSCLC samples
Hematoxylin and eosin (H&E) stained sections were used for morphological evaluation purposes and unstained sections were utilized for immunohistochemical (IHC) staining using standard methods. Briefly, 3–5 µm formalin-fixed, paraffin embedded sections were deparaffinized in 100% xylene for 3 times for 5 min each. Next, all slides were dehydrated in a sequence of ethanol (100%, 70%, 50%) solutions for 3 minutes each. Sections were then subjected to heat-induced epitope retrieval with 0.01 M citrate buffer (pH 6.0).
IHC staining was performed on Ventana Benchmark GX IHC/ISH system (Roche, Basel, Switzerland), using primary MSI2 antibody (Abcam ab76148, Boston, MA, USA) and Mouse and Rabbit Specific HRP/DAB IHC Detection kit–Micro-polymer (Abcam ab236466, Boston, MA, USA) as a detection system. As a negative control, the primary antibody was replaced with normal mouse/rabbit IgG to confirm absence of specific staining. Additionally, all sections were stained using hematoxylin-eosin stain. In the end, all sections were dehydrated by immersion to a sequence of ethanol solutions (70%, 96%, 100%) and xylene for 30 seconds respectively. Coverslips were mounted using Histomount histological mounting medium (HS-103, National Diagnostics, Atlanta, GA, USA).
All slides were viewed on Leica Aperio CS2 scanner (Leica Biosystems, San Diego, CA, USA). MSI2 expression level in tissue was evaluated quantitatively using H-score approach. H-score was calculated as follows: the percentage of cells at each staining intensity level was calculated, and an H-score was assigned and calculated for each slide using the following formula: [1× (% cells 1+) + 2 × (% cells 2+) + 3 × (% cells 3+)] (11). H-score values in the normal tissue ranged from 35 to 77. Median value of H-score of 170 was used as a cutoff for high MSI2 expression. The reason for this cutoff is because values above 170 represent top two quartiles of MSI2 expression H-scores. H-score quantitation was performed using QuPath digital pathology software (QuPath: Open Source Software for Digital Pathology Image Analysis, https://qupath.github.io/), as previously described (12).
TCGA analysis
For The Cancer Genome Atlas (TCGA) analysis, the most recent datasets for NSCLC (as of April, 2020) were accessed and analyzed using tools available at cBioPortal for Cancer Genomics (http://www.cbioportal.org/) (13). Kaplan-Meyer analysis was performed using the KMPLOT tool with automatically determined cutoff function, comparing high and low MSI2 mRNA expression in TCGA samples (http://kmplot.com) (14).
Statistical analysis
For statistical analysis of MSI2 expression (represented as H-score) we used Wilcoxon rank-sum test. The difference was determined as statistically significant in all cases with a P value <0.05. The results were calculated using Microsoft Excel and GraphPad Prism software. The Kaplan-Meier method was used to construct survival curves, as well as to calculate median survival. Difference between the groups was compared using a log-rank test. The correlation coefficients between protein expression and clinical and pathological parameters were estimated using Spearman correlation. Correlation analysis of MSI2 H-scores and clinical and pathological parameters correlations were performed using SPSS 16.0 for Windows (SPSS Inc., Chicago, Illinois, USA). All p values are two-sided, and P<0.05 is determined as statistically significant difference.
Results
NSCLC patient cohort and clinical characteristics
The cohort examined in the current study consists of 40 patients (70% male; 30% female). All patients signed a written consent for the study and processing of tissue samples in an anonymized fashion. The median age was 55 years (mean is 66 years, standard deviation 8.5 years; range, 38 to 75 years). One patient presented with stage I (2.5%), 14 and 15 patients presented with stage II and III (35% and 37.5%, respectively), and 10 patients (25%) had stage IV. All patients received treatment in accordance with the recommendations of the NCCN and did not participate in clinical trials (Table 1). All patient samples were obtained prior to standard of care treatment. Clinical (age, sex, smoking status) and histopathological (disease stage) characteristics (Table 1) had no impact on the patient outcome in terms of overall survival (OS) and progression-free survival (PFS) (P value >0.05) (Table 2). Overall, the cohort represented general population of NSCLC patients.
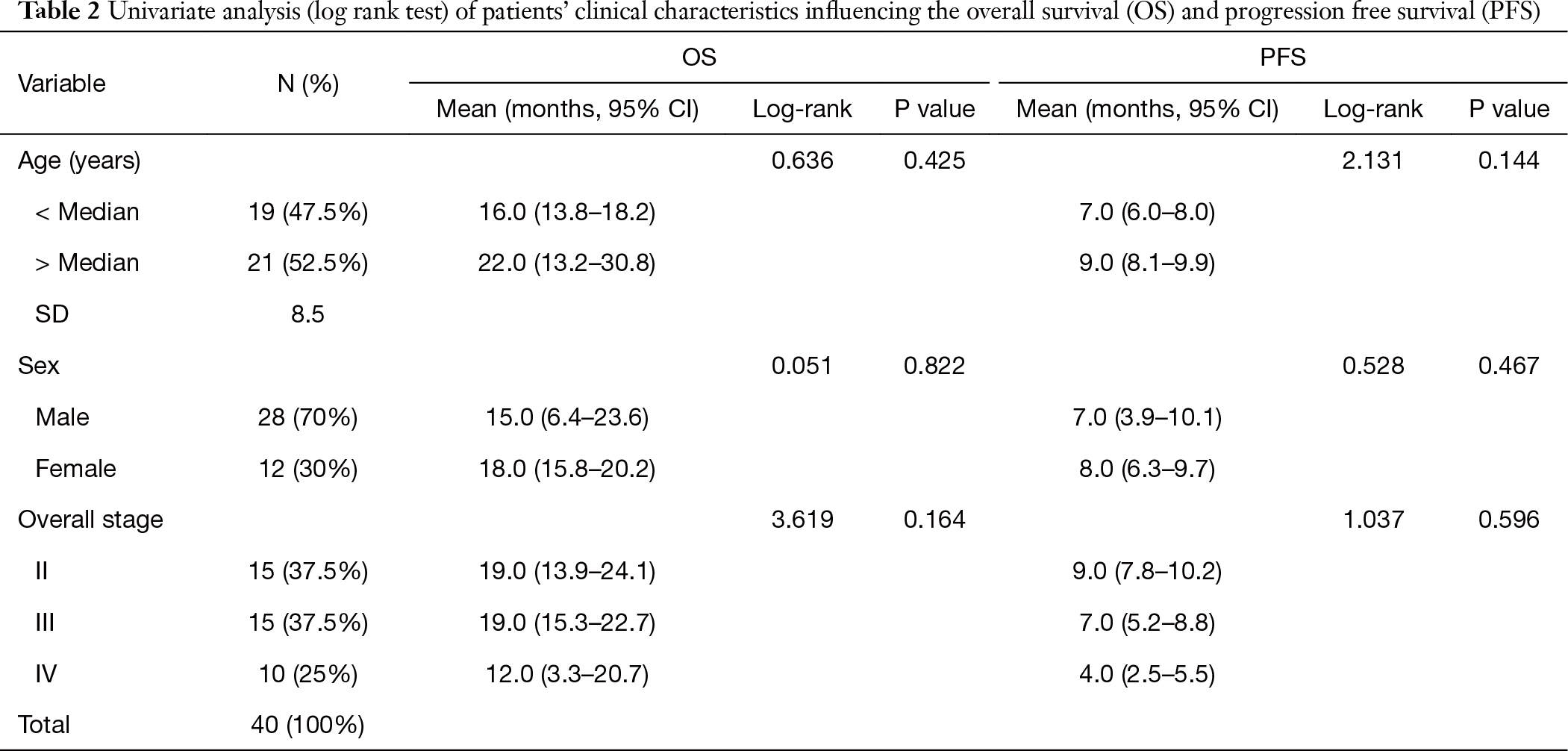
Full table
MSI2 levels are elevated in primary lung tumor tissue versus matching normal lung parenchyma
Based on analysis of lung cancer data in cBioportal database, we performed comparative analysis of MSI2 mRNA expression in a set of normal tissue (n=59) and lung adenocarcinoma samples (n=515) (Figure 1A). The analysis demonstrated, that MSI2 mRNA levels are statistically significantly elevated in primary tumor samples (P<0.0001) compared to normal lung parenchyma. Next, we benchmarked this result to our dataset to reveal, if elevated MSI2 levels in primary lung adenocarcinoma tumors is a feature, represented solely on mRNA level, or it is observed on the protein level as well. Using our patient dataset, we utilized IHC approach to measure expression of MSI2 at the protein level (represented as H-score) in the normal (n=20) and primary tumor (n=40) samples. This allowed us to determine, that MSI2 levels are elevated in primary tumor tissue (median H-score of 170), compared to normal parenchyma as well (P=0.03) (Figure 1B), and therefore we concluded that elevated levels of MSI2 in primary lung adenocarcinoma tumors is a constant feature, both on mRNA and protein levels.
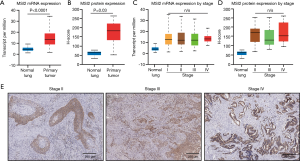
In a subsequent analysis, aiming to determine if MSI2 levels are differentially altered with disease progression, we sorted out samples used in above analysis by disease stage and performed comparative analysis in a similar fashion in between stages. We did not observe statistically significant differences between MSI2 mRNA (Figure 1C) and protein expression (Figure 1D,E) levels depending of disease stage.
Elevated MSI2 levels in primary NSCLC tumors correlate with decreased PFS and OS
IHC approach is one of the most routinely used pathological diagnostics methods, and it can yield substantial data for further patient survival prediction. Kaplan-Meier and correlation analysis of MSI2 expression levels (as determined by IHC) and patient survival in our sample cohort demonstrated, that elevated MSI2 expression (H-score above 170) statistically significantly positively correlated with a decreased PFS combined for all stages (P=0.026). Notably, high levels of MSI2 protein correlated with OS as well, but without statistical difference (P=0.065) (Figure 2A). This assumption is indirectly confirmed by further Kaplan-Meier analysis of MSI2 RNA levels and patients survival using publicly available datasets at cBioportal (14). Results demonstrate, that increased RNA levels of MSI2 in the primary lung tumors positively correlate with a decrease in OS (P=0.02) (Figure 2B), as revealed by survival analysis of TCGA RNA-seq data in NSCLC dataset using Pan-cancer RNA-seq KM plot tool (15). For the other clinical parameters, such as age, gender, smoking, and type of treatment there was no statistically significant correlation for MSI2 expression, both in our cohort of patients and TCGA dataset.
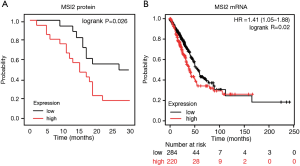
Stage IV NSCLC patients with low MSI2 levels have superior PFS and OS
Next, our goal was to determine, how levels of MSI2 in primary tumors as measured by IHC correlate with NSCLC patients survival during disease progression Univariate log-rank test correlative analysis between MSI2 H-score levels and disease stage in our cohort demonstrated, that there were no statistically significant correlations between increased MSI2 levels on stages II and III and progression-free or OS. However, analysis of stage IV samples revealed, that elevated levels of MSI2 protein (H-scores above median value of 170) statistically significantly correlated with decrease in PFS (P=0.042) and OS (P=0.013) (Table 3, Figure 3A), suggesting that NSCLC patients with low MSI2 levels at a later stage may have better survival.
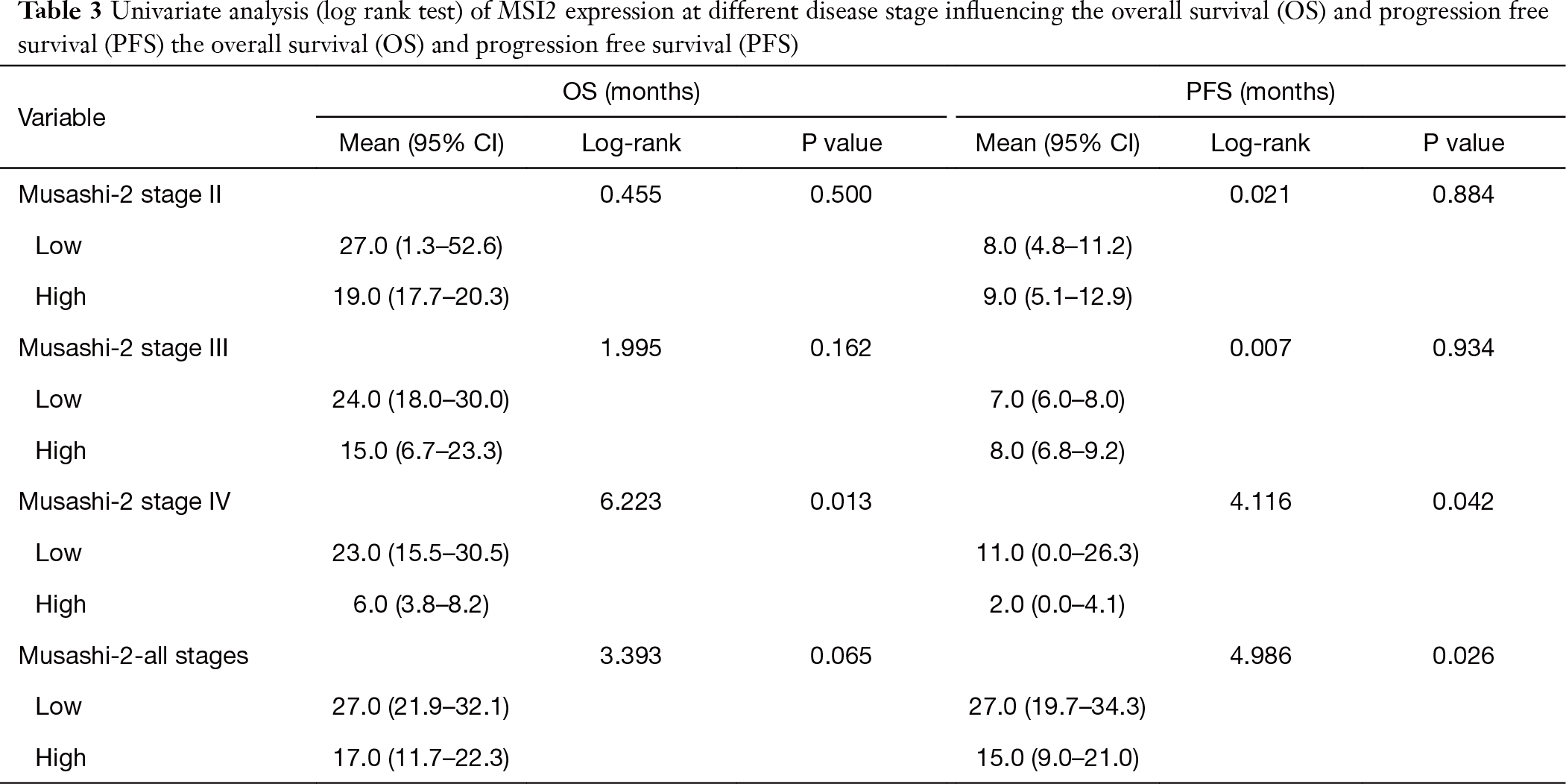
Full table
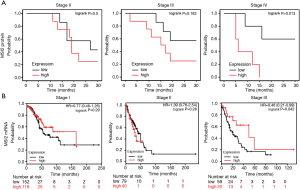
Aiming to determine if the correlation differences (survival and MSI2 protein on stage IV vs. stages II and III) reflect elevated transcription of MSI2 gene or depict enhanced protein activity and/or accumulation, we utilized KM plot tool to examine OS based on high and low levels of mRNA expression in TCGA NSCLC RNA-seq dataset. The limitation of TCGA dataset is that it had less than ten stage 4 NSCLC samples, so we were not able to directly compare our results. TCGA dataset analysis of stage I, II and III NSCLC revealed no statistically significant difference in survival between high and low MSI2 mRNA levels among these stages (Figure 3B), with borderline significance (P=0.042) at stage III.
Discussion
The RNA-binding protein Musashi-2 (MSI2) controls translation of proteins that support stem cell identity and lineage determination (6). Elevated expression of MSI2 plays an important role as a driver of tumor progression (16), dissemination (9), and drug resistance (17,18), in numerous solid (8,19) and hematological malignancies (20-22). In lung cancer settings, previous publications demonstrate that MSI2 expression is elevated from an early stage NSCLC and suggest that MSI2 may play an important role in the reprogramming of transforming growth factor-β (TGF-β) signaling from growth-inhibiting to invasion-promoting during oncogenesis (10).
Despite significant advances in immune-oncology and targeted therapy in the recent years, durable responses in advanced lung cancer only occur in a fraction of patients. Therefore, one of the current challenges is to identify reliable biomarkers to predict therapeutic responses in this patient population. In the current work we for the first time assessed a novel stem cell marker MSI2 as a prediction marker of NSCLC progression. To confirm this assumption, it was necessary to conduct studies not only in the cell lines of NSCLC, but also to perform correlation and IHC analyses of tumor and normal tissue samples obtained from patients with NSCLC.
Our results confirm previously published data on elevated MSI2 levels in primary NSCLC tumors regardless of the stage (10) in an independent patient samples cohort, and for the first time demonstrate that high MSI2 expression levels in primary NSCLC tumors correlate with poor patients survival and prognosis, censored based on three year survival. We observed a statistically significant correlation between high MSI2 expression and lower PFS, but not with OS. This is probably due to relatively small cohort size and can potentially improve and yield statistically more robust values with an increase in number of the samples.
These data can potentially nominate MSI2 to be used as a novel prognostic biomarker in patients with NSCLC. Elevated MSI2 levels in tumor versus normal tissues were detected in extended RNA-seq TCGA dataset, and confirmed in our patients dataset by IHC staining, with comparable results presented. Gene expression profiling by RNA sequencing is now commonly used to profile and classify the cell types present in a tissue, discover new gene signatures for a cell type or pathway, elucidate the developmental lineage of a cell type, and compare different biological states or conditions (23). One advantage of this study is that we used IHC approach to validate RNA-seq findings of gene expression and to evaluate expression of MSI2 in patient samples. Considering the heterogeneity of the tumor, IHC approach allows to evaluate larger area of the tumor and reduce errors in the qualitative and quantitative assessment of expression of the marker of interest, and is one of the most common methods utilized in clinical practice, that can be used to validate the findings from a broad, hypothesis-free approach such as RNA-seq to draw accurate conclusions (24). This, on one hand, can explain demonstrated certain level of discrepancy in results of correlative analysis performed for samples with MSI2 levels measured at protein (via IHC) and mRNA (via RNA-seq) levels. On the other hand, this may reflect underlying intrinsic biological differences, depending on the underlying mechanism of disease progression, when the tumors potentially become more dependent on MSI2 protein activity at later stages of NSCLC oncogenesis.
Conclusions
Based on the clinical and statistical data obtained, we can state a significant role for MSI2 in the processes of carcinogenesis and the progression of the NSCLC. High expression of MSI2 in late-stage tumors and its correlation with decrease in patient survival suggest a higher aggressiveness of the disease in this subset of patients. Measuring MSI2 protein expression in primary NSCLC tumors can be used as a novel potential prognostic biomarker in NSCLC patients. Future studies evaluating MSI2 in an extended patient cohort are warranted.
Acknowledgments
Funding: This work was supported by Russian Foundation for Basic Research project 18-3015-00307 and a William J. Avery Postdoctoral Fellowship from Fox Chase Cancer Center (to AD); by the Russian Government Program for Competitive Growth of KPFU (to AD and YB); by the NCI Core Grant P30 CA006927 to Fox Chase Cancer Center. YB and IT were supported in part by the NIH R21 CA223394, NIH R01 CA218802 grants and by the NCI NIH Core Grant P30 CA060553 to the Robert H Lurie Comprehensive Cancer Center at Northwestern University.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/jtd-20-2787
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jtd-20-2787
Peer Review File: Available at http://dx.doi.org/10.21037/jtd-20-2787
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-20-2787). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by joint institutional ethics board of the Republican Oncological Clinical Hospital, Kazan, Russia and Rostov National Medical Research Center, Rostov-on-Don, Russia (No. 8, 02/13/2018 and No.: 0.1.1.55-16/23/18-8 and). Informed consent was taken from all individual participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Howlader N, Krapcho M, Miller D, et al. SEER Cancer Statistics Review, 1975-2016. Available online: https://seer.cancer.gov/csr/1975_2013/: National Cancer Institute. Bethesda, MD2019.
- Sakakibara S, Nakamura Y, Yoshida T, et al. RNA-binding protein Musashi family: Roles for CNS stem cells and a subpopulation of ependymal cells revealed by targeted disruption and antisense ablation. Proc Natl Acad Sci U S A 2002;99:15194-9. [Crossref] [PubMed]
- Sakakibara S, Nakamura Y, Satoh H, et al. Rna-binding protein Musashi2: developmentally regulated expression in neural precursor cells and subpopulations of neurons in mammalian CNS. J Neurosci 2001;21:8091-107. [Crossref] [PubMed]
- Ito T, Kwon HY, Zimdahl B, et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature 2010;466:765-8. [Crossref] [PubMed]
- Kudinov AE, Karanicolas J, Golemis EA, et al. Musashi RNA-binding proteins as cancer drivers and novel therapeutic targets. Clin Cancer Res 2017;23:2143-53. [Crossref] [PubMed]
- Lee J, An S, Choi YM, et al. Musashi-2 is a novel regulator of paclitaxel sensitivity in ovarian cancer cells. Int J Oncol 2016;49:1945-52. [Crossref] [PubMed]
- Kang MH, Jeong KJ, Kim WY, et al. Musashi RNA-binding protein 2 regulates estrogen receptor 1 function in breast cancer. Oncogene 2017;36:1745-52. [Crossref] [PubMed]
- Guo K, Cui J, Quan M, et al. The Novel KLF4/MSI2 Signaling Pathway Regulates Growth and Metastasis of Pancreatic Cancer. Clin Cancer Res 2017;23:687. [Crossref] [PubMed]
- Kudinov AE, Deneka A, Nikonova AS, et al. Musashi-2 (MSI2) supports TGF-beta signaling and inhibits claudins to promote non-small cell lung cancer (NSCLC) metastasis. Proc Natl Acad Sci U S A 2016;113:6955-60. [Crossref] [PubMed]
- Hirsch FR, Varella-Garcia M, Fau-Bunn PA Jr, et al. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol 2003;21:3798-807. [Crossref] [PubMed]
- Bankhead P, Loughrey MB, Fernández JA, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep 2017;7:16878. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Győrffy B, Surowiak P, Budczies J, et al. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One 2013;8:e82241 [Crossref] [PubMed]
- Nagy Á, Lanczky A, Menyhart O, et al. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep 2018;8:9227. [Crossref] [PubMed]
- Barbouti A, Hoglund M, Johansson B, et al. A novel gene, MSI2, encoding a putative RNA-binding protein is recurrently rearranged at disease progression of chronic myeloid leukemia and forms a fusion gene with HOXA9 as a result of the cryptic t(7;17)(p15;q23). Cancer Res 2003;63:1202-6. [PubMed]
- Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene 2010;29:4741-51. [Crossref] [PubMed]
- Fang T, Lv H, Wu F, et al. Musashi 2 contributes to the stemness and chemoresistance of liver cancer stem cells via LIN28A activation. Cancer Lett 2017;384:50-9. [Crossref] [PubMed]
- Yang C, Zhang W, Wang L, et al. Musashi-2 promotes migration and invasion in bladder cancer via activation of the JAK2/STAT3 pathway. Lab Invest 2016;96:950-8. [Crossref] [PubMed]
- Aly RM, Ghazy HF. Prognostic significance of MSI2 predicts unfavorable outcome in adult B-acute lymphoblastic leukemia. Int J Lab Hematol 2015;37:272-8. [Crossref] [PubMed]
- Byers RJ, Currie T, Tholouli E, et al. MSI2 protein expression predicts unfavorable outcome in acute myeloid leukemia. Blood 2011;118:2857-67. [Crossref] [PubMed]
- Thol F, Winschel C, Sonntag AK, et al. Prognostic significance of expression levels of stem cell regulators MSI2 and NUMB in acute myeloid leukemia. Ann Hematol 2013;92:315-23. [Crossref] [PubMed]
- Chu Y, Corey DR. RNA sequencing: platform selection, experimental design, and data interpretation. Nucleic Acid Ther 2012;22:271-4. [Crossref] [PubMed]
- Byron SA, Van Keuren-Jensen KR, Engelthaler DM, et al. Translating RNA sequencing into clinical diagnostics: opportunities and challenges. Nat Rev Genetics 2016;17:257-71. [Crossref] [PubMed]


