
Abnormal spindle-like microcephaly-associated protein enhances cell invasion through Wnt/β-catenin-dependent regulation of epithelial-mesenchymal transition in non-small cell lung cancer cells
Introduction
Lung cancer is the leading cause of cancer mortality in both men and women. It contributes to 18.4% of total cancer deaths, as indicated by the GLOBOCAN 2018 global cancer statistics (1). Conventional chemotherapy for non-small-cell carcinoma (NSCLC) has generally included platinum-based treatment for more than 3 decades (2), which has been associated with side-effects. In contrast, the use of representative inhibitors of epidermal growth factor receptor and anaplastic lymphoma kinase as targeted treatment for lung cancer has been shown to overcome the shortcomings of conventional therapy and lead to a favorable prognosis (3-8). Clinical trials of human programmed cell death 1 (PD-1) antibodies have been demonstrated prolonged overall survival in patients with advanced NSCLC (9-13). However, with chemoresistance and metastasis still a challenge, novel targets for effective treatments of advanced lung cancer are urgently required.
Abnormal spindle-like microcephaly-associated protein (ASPM) is the human ortholog of the Drosophila ASP and a centrosomal protein normally regulating neural development and brain size (14). The overexpression of ASPM had been identified in multiple types of cancer, such as glioblastoma, hepatocellular carcinoma (HCC), prostate cancer, and pancreatic cancer (15,16). ASPM enhances the stemness and progression of various cancers via the activation of the Wnt/β-catenin signaling pathway (17-21).
Of note, there is no obvious link between the ASPM gene and epithelial-mesenchymal transition (EMT)-mediated invasion in lung cancer studies, particularly in the prevalent NSCLC subtypes of cancer. Most studies on ASPM in lung cancer are restricted to the bioinformatics field. In one study, differentially expressed genes were screened from The Cancer Genome Atlas database, and 14 targeted genes, including ASPM, were found to be associated with patient survival (22). New druggable targets confirmed ASPM as a promising target in a breast cancer cell line model and lung cancer cell line model (23). ASPM was significantly upregulated in patients with different lung cancer subtypes (24). To identify better targeted treatments for small-cell lung cancer patients, exome sequencing was performed to screen genes frequently mutated; 8 of 36 genes, including ASPM, were found (25). However, none of these studies validated the mRNA and protein expression levels of ASPM. To the best of our knowledge, the present study was the first to investigate the functional role of ASPM in relation to EMT-induced cell invasion in lung cancer. It was found that the silencing of ASPM strongly attenuated invasion. Mechanistically, it was demonstrated that the ectopic expression of ASPM increased cell invasion through the regulation of the EMT process or matrix metalloproteinases (MMPs). The silencing of ASPM significantly reduced EMT markers and MMP protein levels. Rescue experiments suggested that ASPM enhanced EMT-mediated invasion via the Wnt/β-catenin pathway in NSCLC. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/jtd-21-566).
Methods
Patient samples
All patient samples were obtained from the specimen bank of the central laboratory of the Affiliated Brain Hospital of Nanjing Medical University (Nanjing, China). All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The present study was approved by the ethics committee of the Affiliated Brain Hospital of Nanjing Medical University (No. 20180407). Written informed consent was obtained all patients.
GEO data mining and protein-protein interaction (PPI) networks
The GSE19804, GSE18842, and GSE33532 datasets were downloaded from the microarray GEO datasets and analyzed (26-29). Adjusted P<0.01 and log fold change (FC) >1.5 were considered statistically significant. The most significant differentially expressed genes were enriched for PPI via STRING (https://string-db.org/) and Cytoscape software (https://cytoscape.org/).
Cell culture
Human lung cancer A549 and Calu-3 cells and human normal alveolar BEAS-2B cells were purchased from the Chinese Academy of Science Committees Type Culture Collection Cell Bank (Shanghai, China). The authenticity of these cell lines was confirmed by the Chinese Academy of Science Committee Type Culture Collection Cell Bank before purchase by STR DNA typing methodology. All cell lines were grown in RPMI-1640 medium (Thermo Fisher Scientific, Shanghai, China) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Thermo Fisher Scientific, Shanghai, China) and penicillin-streptomycin solution (80 µ/mL penicillin and 80 µg/mL streptomycin; Nanjing KeyGen Biotech). All cells were cultured in a humidified incubator containing 5% CO2 at 37 °C, unless otherwise stated.
Gene knockdown and lentiviral transfection
Silencing of ASPM cells was generated using the small interference RNA (siRNA) duplex against human ASPM: si-ASPM, 5'-UGCCAUGGUGCAACUUGCU-3' (Guangzhou RiboBio). The siRNA negative control (si-NC) 5'-UUACCUCUAGUCGUCAUGU-3' (scrambled) was used as an NC. ASPM siRNA was transfected into A549 and Calu-3 cells by Lipofectamine 2000 (Thermo Fisher Scientific), according to the manufacturer’s instructions. Lentivirus-mediated ASPM overexpression or NC vectors were purchased from Shanghai GeneChem. When 293T cells, genetically modified to express green fluorescent protein (GFP), reached 60% confluence, they were packaged with 3rd-generation lentiviral vectors for 2 rounds in a row containing pHelper 1.0 vector and pHelper 2.0 vector plasmid; virus supernatants were harvested every 24 h. The day before transfection, lung cancer cells were plated in a 6-well plate at 60% confluence in a medium without FBS or antibiotics. The virus supernatant was infected with lung cancer cell lines using Lipofectamine 2000 when they reached 80% confluence twice at 36 and 48 h, in order to reach the highest infection efficiency. Polybrene (hexadimethrine bromide, 5 µg/µL), which can increase transfection efficiency) was also used in the transfection procedure to increase infection efficiency. The previous supernatant was discarded, and serum containing growth medium was added at the time of the second infection. The cells stably expressed ASPM were selected using puromycin antibiotics for more than one week and expanded for further study.
RNA isolation and quantitative reverse transcription quantitative polymerase chain reaction (qRT-PCR)
Total RNA was isolated from lung cancer A549 and Calu-3 cell lines using TRIzol Reagent (Beijing Solarbio Science & Technology), according to the manufacturer’s instructions. Lysis buffer (300 µL) was added to 10–20 mg tumor tissues, followed by homogenization with 10 µK Proteinase K diluted in 590 µL RNase-free water. The sample mixture was incubated for 10–20 min at 56 °C, and absorption columns were applied to each sample according to the manufacturer’s instructions (Tiangen Biotech). RNA concentration was measured using an ultramicro-ultraviolet spectrophotometer (SMA2000; Thermo Fisher Scientific). Total RNA (2 µg) was converted to cDNA using the Superscript II One-Step RT-PCR System (Yeasen Biotechnology [Shanghai]), according to the manufacturer’s instructions. cDNA was transcribed and synthesized using PrimeScript RT Reagent Kit (Takara Bio) as follows: priming at 25 °C for 5 min, reverse transcription at 37 °C for 30 min, and reverse transcription inactivation at 85 °C for 5 min. The qRT-PCR Master Mix (Tiangen Biotech) was used for qRT-PCR in a 20-µL reaction system containing 1 µL cDNA, 10 µL SuperReal PreMix Plus, 0.5 µL forward and reverse primer of ASPM at a concentration of 10 µM, and nuclease-free water. qRT-PCR was performed at 95 °C for 3 min (pre-denaturation), followed by 40 cycles of 95 °C for 15 s (denaturation) and 60 °C for 45 s (annealing and extension), and a single cycle of 95 °C for 15 s, 60 °C for 60 s, and 95 °C for 15 s to generate dissociation curves (FQD-96A; Hangzhou Bori Technology). The GAPDH gene was used as an endogenous control. The primer sequences used were as follows: GAPDH, 5'-AATCCCATCACCATCTTCCA-3' forward and 5'-TGGACTCCACGACGTACTCA-3' reverse; β-catenin si-RNA, 5'-CGGAGGAGAUGUACAUUCAdTdT-3'; β-catenin si-NC, 5'-UUCUCCGAACGUGUCACGUdTdT-3'. All PCR reactions were performed in triplicate, and the specificity of the reaction was determined by melting curve analysis. Comparative quantification of each target gene was performed based on cycle threshold normalized to GAPDH using the ΔΔCq method to calculate the relative expression level. Three separate experiments were repeated for each cell line. Expression differences were analyzed by 2-tailed Student’s t-tests.
Western blot analysis
The transfected cells were harvested and lysed on ice in radioimmunoprecipitation assay buffer [20 mM Tris-HCl (pH 7.5)], 150 mM NaCl, 1 mM Na2 ethylenediaminetetraacetic acid, 1 mM EGTA, 1% NP-40, 1% sodium deoxycholate, 2.5 mM sodium pyrophosphate, 1 mM beta-glycerophosphate, 1 mM Na3VO4, 1 µg/mL leupeptin; Bei Si Te Biotechnology) for 15 min containing the protease inhibitor phenylmethylsulfonyl fluoride (Bei Si Te Biotechnology). Subsequently, the protein samples were centrifuged at 16,099 ×g for 15 min and heated at 100 °C for a further 10 min. The concentration of quantified protein lysates was determined by the BCA protein assay (Beyotime Institute of Biotechnology), and equal amounts of protein (50 µg) were loaded onto gels, separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and then transferred to polyvinylidene fluoride membranes (EMD Millipore). The membranes were washed 3 times with TBST [20 mM Tris-HCl, 150 mM NaCl, 0.1% (v/v) Tween 20] and 0.1% Tween-20 (Yi Fei Xue Biotechnology). Next, the membranes were blocked with 5% non-fat milk for 1 h and incubated with primary antibodies, according to the manufacturer’s instructions, at 4 °C overnight, followed by a 1.5-h incubation with horseradish peroxidase-conjugated secondary antibodies (anti-rabbit or anti-mouse immunoglobulin G; dilution: 1:2,000). The protein bands were visualized by enhanced chemiluminescence assay (Yeasen Biotechnology [Shanghai].) and observed using Image Lab software and the Odyssey CLx system (Gene Company Limited). GAPDH was used as the internal standard. The primary antibodies used were as follows: E-cadherin (dilution: 1:1,000; ProteinTech Group), MMP2 (dilution: 1:500; Abcam), MMP9 (dilution: 1:1,000; Abcam), Glycogen synthase kinase-3β (Gsk-3β) (dilution: 1:2,000; Cell Signaling Technology), β-catenin (dilution: 1:2,000; Abcam), and GAPDH (1:10,000; Abcam).
Immunofluorescence staining assay
Lung cancer cells were fixed with 4% formaldehyde (Sinopharm Chemical Reagent) for 1 h, and perforated with 0.1% Triton X (Beijing Solarbio Science & Technology) for 0.5 h. The sections were then incubated with 5% bovine serum albumin (BSA) at room temperature for 5 min, and with E-cadherin, N-cadherin, or ASPM primary antibody (Thermo Fisher Scientific) at room temperature for 2 h. Sections were then incubated with a secondary antibody (Cervico Biological Technology, China) at room temperature for 1 h. Subsequently, they were incubated with 4’,6’-diamidino-2-phenylindole dihydrochloride (DAPI; Bei Si Te Biotechnology) at room temperature for 10 min. Images for all conditions were obtained using identical acquisition parameters, and fluorescence intensity was analyzed using ImageJ software (National Institutes of Health).
Wound healing assay
A wound healing assay was performed on A549 and Calu-3 cells transfected with siRNA against ASPM or lentivirus-mediated ASPM (Lv-ASPM). The monolayer of cells at 80–90% confluence was scratched with a 200-µL pipette tip and washed twice with phosphate-buffered saline (PBS) to remove cell debris and floating cells. Multiple views of the leading edge of the scratch were photographed under an inverted microscope at the following 3 time points: 0, 24, and 48 h. Three independent experiments were performed and quantified using ImageJ software .
Invasion assay
The invasion assay was carried out using a 24-well Boyden’s chamber. Lung cancer cells (200 µL; 1×105), A549 and Calu-3, were seeded into the upper chambers, which were coated with Matrigel (8-µm pore size; BD Biosciences). The chambers were then inserted into a Transwell apparatus (Corning). Cells in the upper chamber were cultured in serum-free RPMI-1640 medium. RPMI-1640 medium (600 µL) with 10% fetal bovine serum was added to the lower chamber. The chamber was incubated for 48 h at 37 °C in 5% CO2. The cells on the upper surface were removed by cotton swab. The bottom of the inserts was rinsed with 1× PBS and fixed in 80% methanol, followed by staining with 1% crystal violet (Google Biotechnology) for 15 min. The invaded cells were then counted under a microscope at high-power fields (Olympus) and analyzed by 2-tailed Student’s t-tests. Each experiment was repeated at least 3 times.
Statistical analysis
Data from at least three independent experiments are presented as the mean ± standard error of the mean or as the mean ± standard deviation, where n=3 (3 independent experiments performed with three different cell generations), unless otherwise stated. Statistical analysis was performed using GraphPad Prism 7.0 (GraphPad Software). Measurement data analysis between two groups was performed using a 2-tailed Student’s t-test. One-way analysis of variance with Bonferroni correction was used to evaluate the differences among more than three groups. P<0.05 indicated a statistically significant difference.
Results
Screening the most significant differentially expressed genes and generation of the ASPM-knockdown and -overexpression lung cancer models
The GSE19804, GSE18842, and GSE33532 datasets were downloaded from the National Center of Biotechnology information (NCBI) website. In total, 46 lung cancer and 45 normal tissues were included in the GSE18842 dataset, 60 paired lung cancer and normal tissues were included in the GSE19804 dataset, and paired lung cancer and normal tissues with 20 samples for each group were found in the GSE33532 dataset. Differentially expressed genes between NSCLC and normal lung tissues were selected using GEO2R software (https://www.ncbi.nlm.nih.gov/geo/geo2r/) with the following criteria: adjusted P<0.01 and absolute value of logFC >1.5. In total, 355 overlapping genes were identified as significantly changed candidate genes among the 3 datasets generated by the Venn diagram (Figure 1A). Subsequently, the 355 differentially expressed genes were analyzed by STRING. The PPI networks created involved 346 nodes and 1,119 edges (Figure 1B). To further screen the most significant differentially expressed genes, we used MCODE mode in Cytoscape. The criteria for selection were MCODE scores >5, degree cut-off =2, node score cut-off =0.2, maximum depth =100, and k-score =2. Based on the selection criteria, 26 nodes and 309 edges were selected and 26 genes were considered as the most enriched genes, including the ASPM molecule (Figure 1C). To validate whether ASPM plays an indispensable role in lung cancer cells, we compared the differential expression levels of ASPM between lung cancer and normal tissues; ASPM protein expression was detected using pair-matched normal, lung cancer, and cancer-adjacent tissues. A higher level of ASPM expression was identified in lung cancer tissues compared with their counterpart cancer-adjacent tissues. The ASPM expression has a minimal effect on normal tissue (Figure 1D). ASPM was successfully identified as the most differentially upregulated gene in lung cancer tissues. Next, the mRNA expression of ASPM was significantly higher in both A549 and Calu-3 cell lines compared with the normal alveolar BEAS-2B cells (Figure 1E). ASPM protein analysis was consistent with our previous mRNA expression in both NSCLC cell lines, with lung cancer cell lines exhibiting higher levels of ASPM than the control cells (Figure 1F). The transient knockdown of ASPM was achieved using si-ASPM in both A549 and Calu-3 cell lines. Immunofluorescence microscopy demonstrated successful elimination of ASPM in both lung cancer cell lines due to the siRNA vector containing GFP, which could be observed under the fluorescent field (Figure 1G). Transfection efficiency was relatively higher than in the NC group, which was transfected with an empty scrambled vector (NC). In addition, Lv-ASPM was used to overexpress ASPM in both A549 and Calu-3 cell lines. Transfection efficiency was again found to be higher in both lung cancer cell lines than in their counterpart NC groups (Figure 1G). The mRNA expression of ASPM was analyzed in both A549 and Calu-3 lung cancer cell lines, with the si-ASPM expression markedly reduced compared with the si-NC group, and the Lv-ASPM (overexpression of ASPM) mRNA levels significantly elevated compared with the Lv-NC groups in both lung cancer cell lines (P<0.001) (Figure 1H). The knockdown and overexpression of ASPM in a lung cancer cell line model was successfully generated and used in subsequent experiments.
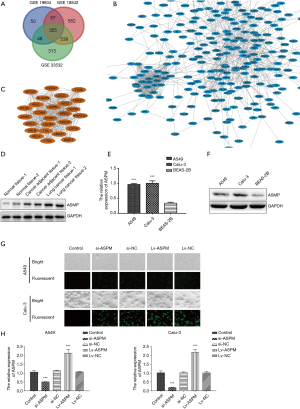
ASPM enhances cell invasion in lung cancer
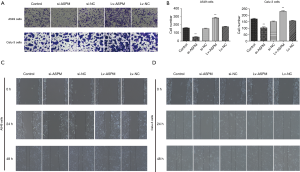
To evaluate whether ASPM plays an indispensable role in cell invasion in both A549 and Calu-3 cell lines, invasiveness was compared among the control, ASPM-silencing, and -overexpression groups. The ectopic expression of ASPM markedly upregulated cell invasion compared with the empty control groups in both A549 and Calu-3 cell lines (Figure 2A,B). ASPM-knockdown prominently decreased the number of invaded lung cancer cells compared with the control groups (Figure 2A,B); quantification data are presented in Figure 2B (si-ASPM vs. si-NC and Lv-ASPM vs. Lv-NC, P<0.01). The wound healing assay demonstrated that the silencing of ASPM significantly decreased cancer cell motility in a time-dependent manner, while the ectopic expression of ASPM closed the wound faster than the control group in both lung cancer cell lines (Figure 2C,D). ASPM enhanced cell invasion and migration in both lung cancer cell lines.
ASPM enhances invasion through the regulation of EMT and MMP proteins
Next, the underlying mechanism of ASPM-mediated invasion was investigated. EMT is a common pathway for the invasion of tumor cells into the surrounding tissues (30). Therefore, the mRNA expression of typical epithelial and mesenchymal markers was analyzed. N-cadherin and vimentin are representative mesenchymal markers (Figure 3). Their mRNA and protein levels were both markedly upregulated in ASPM-overexpressing (Lv-ASPM) A549 and Calu-3 lung cancer cells (Lv-ASPM vs. Lv-NC, P<0.01), but significantly downregulated in ASPM-lacking (si-ASPM) cancer cells (si-ASPM vs. si-NC, P<0.01) (Figure 3A,B,C,D). In contrast, E-cadherin, an epithelial marker, was reduced in its mRNA and protein expressions in both lung cancer cell lines. Snail is an EMT-related transcription factor, and its expression was increased in ASPM-overexpressing A549 and Calu-3 cells (Figure 3A,B,C,D, P<0.01). Immunofluorescence also confirmed the Western blot analysis results, namely that E-cadherin expression was increased in the si-ASPM groups in both lung cancer cell lines, while N-cadherin was mainly expressed in the Lv-ASPM groups in both A549 and Calu-3 lung cancer cell lines (Figure 3G,H).
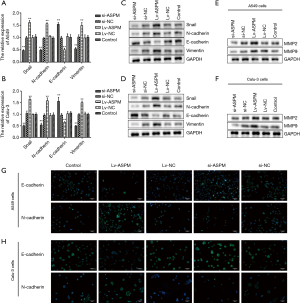
Another possible pathway that tumor cells hijacked is the secretion of a certain amount of MMPs in order to clear a path for a bulk of cancer cells to invade. MMP2 and MMP9 increased when ASPM was successfully overexpressed in a A549 and Calu-3 cell line model. It was also observed that the transient silencing of ASPM caused a reduction in the MMP2 and MMP9 protein levels (Figure 3E,F). ASPM promoted invasion through the regulation of E-cadherin, vimentin, and Snail, or MMP proteins.
ASPM affects EMT-mediated invasion via the Wnt/β-catenin signaling pathway
To determine the mechanism of signaling pathways in the EMT-mediated invasion process, the Wnt/β-catenin signaling pathway was explored, due to its correlation with EMT in cancer and the ability of β-catenin to form complexes with E-cadherin (31) (Figure 4). As expected, the mRNA and protein levels of Wnt-3a and β-catenin were reduced when lung cancer cells lacked ASPM expression (si-ASPM) (Figure 4A,B,C,D). However, the overexpression of ASPM (Lv-ASPM) led to a higher expression of Wnt-3a and β-catenin in both A549 and Calu-3 cancer cell lines (Figure 4A,C). Gsk-3β mRNA expression had the opposite effect to that of β-catenin when the ASPM gene expression was manipulated in both lung cancer cell lines (Figure 4A,C). The downregulation of ASPM caused an increase in Gsk-3β protein levels, and the overexpression of ASPM reduced those levels; data obtained in both lung cancer cell lines (A549 and Calu-3) were consistent (Figure 4B,D). Gsk-3β was supposed to form a complex with β-catenin when the Wnt/β-catenin signaling pathway was inactivated. Once Gsk-3β was activated and phosphorylated, this complex dissociated β-catenin, due to the ubiquitination that helped degrade it. It was observed that Gsk-3β protein levels were negatively associated with the expression of β-catenin, either through the silencing or ectopic expression of ASPM, in both A549 and Calu-3 cancer cell lines (Figure 4B,D). The phosphorylated form of Gsk-3β (p-Gsk-3β) had a similar trend as that of β-catenin molecules in relation to protein levels, when ASPM levels were genetically altered. For example, the overexpression of ASPM increased the protein levels of β-catenin and p-Gsk-3β (Figure 4B,D). However, the silencing of ASPM reduced the expression of β-catenin and p-Gsk-3β (Figure 4B,D). This suggested that ASPM affects invasion through Wnt/β-catenin signaling transduction.
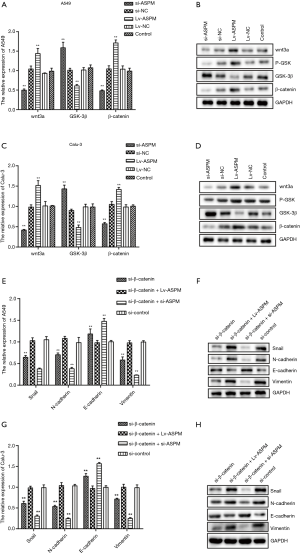
To understand whether ASPM promotes EMT-mediated invasion via the Wnt/β-catenin signaling pathway, the inhibition of β-catenin was induced to check whether any of EMT-related molecules changed accordingly. It was found that the mRNA level of E-cadherin increased, but that of all other mesenchymal markers (N-cadherin and vimentin) or Snail decreased (Figure 4E,G). Blockage of β-catenin decreased the protein levels of Snail, N-cadherin, and vimentin. However, when ASPM was overexpressed and β-catenin inhibited at the same time in both A549 and Calu-3 cancer cell lines, Snail, N-cadherin, and vimentin protein levels reverted to normal levels, as observed in our control group (Figure 4F,H). This suggested that ASPM affected EMT markers and Snail through the Wnt/β-catenin signaling pathway. The silencing of both β-catenin and ASPM significantly reduced the protein levels of Snail, N-cadherin, and vimentin in both A549 and Calu-3 cell line models (Figure 4F,H). The overexpression of ASPM impaired the E-cadherin protein expression in the 2 lung cancer cell lines with the transient silencing of the β-catenin molecule. The silencing of ASPM in the transient knockdown of β-catenin cancer cells (both A549 and Calu-3) can restore the protein levels of E-cadherin (Figure 4F,H). Overall, the Wnt/β-catenin signaling pathway plays an indispensable role in cancer cell invasion by affecting the EMT process. ASPM promoted EMT-mediated invasion through the Wnt/β-catenin signaling pathway in lung cancer cells.
Discussion
Lung cancer is the most commonly diagnosed cancer and the leading cause of cancer mortality worldwide (1). NSCLC accounts for 80–85% of all lung cancer cases (32). Targeted therapy and clinical trials of PD-1 antibodies have been shown to have better clinical benefits than conventional therapy and have been linked to a longer overall survival rate in advanced NSCLC patients (2,3). However, the 5-year overall survival rate of patients with metastatic lung cancer is still poor. It is therefore necessary to develop new targets for patients with advanced lung cancer.
ASPM was initially identified as a centrosomal protein modulating mitotic spindle regulation and neural development (33). HCC with ASPM overexpression was associated with a lower 5-year survival than HCC with a low ASPM expression (18). The microarray data of ASPM was validated by qRT-PCR, and ASPM upregulation was observed in highly invasive glioblastoma multiforme compared with benign pilocytic astrocytomas (34). The higher expression of ASPM is associated with advanced tumor grade in breast cancer (35). ASPM has been verified as a molecular tissue marker for early HCC (17,18). Consistent with the above findings, we analyzed the differential expressions of ASPM among lung cancer tissues, normal tissues, and lung cancer cells (Figure 1A,B). The increase in ASPM expression was validated in the lung cancer tissues and NSCLC cells in the present study (Figure 1A,B). Another previously published study also reported that the expression of ASPM in lung cancer was significantly positively correlated with poor prognosis in 90 tumor and benign pulmonary lesions (36). ASPM has been identified as a differentially expressed gene using GEO data mining in NSCLC (37). However, validation experiments to elucidate the functional role of ASPM are still lacking. Our research validates the functional role of ASPM-mediating cell migration and invasion in NSCLC cells (Figure 2). However, mechanism of how ASPM promotes invasion in human lung cancer has not been fully investigated. EMT has been shown to trigger the dissociation of cancer cells from primary carcinomas, those cancer cells subsequently disseminated to the distant metastatic sites (38). The silencing of Sirtuin 6 inhibited transforming growth factor β1-induced EMT to decrease invasion in NSCLC (39). Early tumor recurrence was twice as likely to occur in HCC with ASPM overexpression than in HCC with low ASPM expression (18). The findings of the present study indicate that the protein levels of MMP2 and MMP9 were reduced when ASPM was reduced in both NSCLC A549 and Calu-3 cell lines. The increased expression of ASPM facilitated MMP2/9 induction (Figure 3E,F). NSCLC patients with a high expression of forkhead box P3 (FOXP3) exhibited a significant decrease in overall and recurrence-free survival rates. The ectopic expression of ASPM increased Slug, Snail and MMP9 protein expression. This suggests that FOXP3 induces Wnt/β-catenin signaling and upregulates Wnt downstream target genes (40). The findings of the present study indicated that ASPM favored Snail. If FOXP3 can independently affect Snail and MMP9, it is likely that ASPM regulates MMP9 protein expression via FOXP3. Clinical correlative studies revealed that ASPM promotes prostate and pancreatic cancer stemness and progression through the upregulation of Wnt/β-catenin signaling (20). The silencing of ASPM results in reduced Wnt-mediated transcription. The co-expression of stabilized β-catenin and Wnt rescued brain injury with the in vivo knockdown of ASPM in the developing brain (21). A recent study noted that ASPM interacts with disheveled-3, the upstream regulator of canonical Wnt signaling to avoid β-catenin degradation in prostate cancer cells (19). It was also observed that the lack of ASPM reduced the expression of N-cadherin, vimentin, and Snail which rendered the lung cancer cells with a lesser mesenchymal phenotype (Figure 3). Increased levels of ASPM significantly decreased E-cadherin expression, but increased that of vimentin and Snail (Figure 3). The present findings strengthened other studies’ results that ASPM is the upstream regulator of Wnt/β-catenin signaling (Figure 4). The ratio of p-GSK-3β to GSK-3β in A549 lung cancer cells among different groups were as follows: Lv-NC: 0.53, Lv-ASPM: 2.8, si-NC: 0.67, and si-ASPM: 0.13. The ratio of p-GSK-3β to GSK-3β in Calu-3 lung cancer cells among different groups were as follows: Lv-NC: 1.08, Lv-ASPM: 3.92, si-NC: 0.89, and si-ASPM: 0.33. The overexpression of ASPM combined with the blockage of β-catenin successfully restored mRNA and protein levels of N-cadherin, vimentin, and Snail during the EMT process (Figure 4). In summary, our novelty rely on that ASPM promoted EMT-mediated cells invasion via the Wnt/β-catenin signaling pathway in NSCLC cells. However, our research still has the potential to improve in the future, for example, independent clones of si- ASPM will be included in our following study, as our current study only contained two siRNA vectors in two cell lines model. To further explore the Wnt downstream effectors, we will test Axin2, Myc, and cyclin D1 in a future study.
In conclusion, we found that ASPM enhances NSCLC invasion through EMT markers and by affecting MMP2/9 proteins. Wnt/β-catenin signaling is required for ASPM-induced EMT-mediated invasion in lung cancer.
Acknowledgments
Funding: Funding was provided by Nanjing Medical University (grant No. JX102GSP201727).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/jtd-21-566
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jtd-21-566
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-21-566). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The present study was approved by the by the ethics committee of the Affiliated Brain Hospital of Nanjing Medical University (No. 20180407). Written informed consent was obtained all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. Erratum in: CA Cancer J Clin 2020;70:313. [Crossref] [PubMed]
- Sabari JK, Lok BH, Laird JH, et al. Unravelling the biology of SCLC: implications for therapy. Nat Rev Clin Oncol 2017;14:549-61. [Crossref] [PubMed]
- Schiller JH, Harrington D, Belani CP, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med 2002;346:92-8. [Crossref] [PubMed]
- Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380-8. [Crossref] [PubMed]
- Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. [Crossref] [PubMed]
- Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. [Crossref] [PubMed]
- Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010;363:1693-703. [Crossref] [PubMed]
- Solomon BJ, Besse B, Bauer TM, et al. Lorlatinib in patients with ALK-positive non-small-cell lung cancer: results from a global phase 2 study. Lancet Oncol 2018;19:1654-67. [Crossref] [PubMed]
- Mok TS, Wu YL, Kudaba I, et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet 2019;393:1819-30. [Crossref] [PubMed]
- Herbst RS, Baas P, Kim DW, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 2016;387:1540-50. [Crossref] [PubMed]
- Horn L, Mansfield AS, Szczesna A, et al. First-Line Atezolizumab plus Chemotherapy in Extensive-Stage Small-Cell Lung Cancer. N Engl J Med 2018;379:2220-9. [Crossref] [PubMed]
- Ready N, Farago AF, de Braud F, et al. Third-Line Nivolumab Monotherapy in Recurrent SCLC: CheckMate 032. J Thorac Oncol 2019;14:237-44. [Crossref] [PubMed]
- Rudin CM, Poirier JT, Byers LA, et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer 2019;19:289-97. [Crossref] [PubMed]
- Williams SE, Garcia I, Crowther AJ, et al. Aspm sustains postnatal cerebellar neurogenesis and medulloblastoma growth in mice. Development 2015;142:3921-32. [Crossref] [PubMed]
- Horvath S, Zhang B, Carlson M, et al. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc Natl Acad Sci U S A 2006;103:17402-7. [Crossref] [PubMed]
- Peng L, Liu Z, Xiao J, et al. MicroRNA-148a suppresses epithelial-mesenchymal transition and invasion of pancreatic cancer cells by targeting Wnt10b and inhibiting the Wnt/beta-catenin signaling pathway. Oncol Rep 2017;38:301-8. [Crossref] [PubMed]
- Wurmbach E, Chen YB, Khitrov G, et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology 2007;45:938-47. [Crossref] [PubMed]
- Lin SY, Pan HW, Liu SH, et al. ASPM is a novel marker for vascular invasion, early recurrence, and poor prognosis of hepatocellular carcinoma. Clin Cancer Res 2008;14:4814-20. [Crossref] [PubMed]
- Pai VC, Hsu CC, Chan TS, et al. ASPM promotes prostate cancer stemness and progression by augmenting Wnt-Dvl-3-beta-catenin signaling. Oncogene 2019;38:1340-53. [Crossref] [PubMed]
- Wang WY, Hsu CC, Wang TY, et al. A gene expression signature of epithelial tubulogenesis and a role for ASPM in pancreatic tumor progression. Gastroenterology 2013;145:1110-20. [Crossref] [PubMed]
- Buchman JJ, Durak O, Tsai LH. ASPM regulates Wnt signaling pathway activity in the developing brain. Genes Dev 2011;25:1909-14. [Crossref] [PubMed]
- Li L, Peng M, Xue W, et al. Integrated analysis of dysregulated long non-coding RNAs/microRNAs/mRNAs in metastasis of lung adenocarcinoma. J Transl Med 2018;16:372. [Crossref] [PubMed]
- Cheng F, Zhao J, Fooksa M, et al. A network-based drug repositioning infrastructure for precision cancer medicine through targeting significantly mutated genes in the human cancer genomes. J Am Med Inform Assoc 2016;23:681-91. [Crossref] [PubMed]
- Zhang Y, Wang H, Wang J, et al. Global analysis of chromosome 1 genes among patients with lung adenocarcinoma, squamous carcinoma, large-cell carcinoma, small-cell carcinoma, or non-cancer. Cancer Metastasis Rev 2015;34:249-64. [Crossref] [PubMed]
- Iwakawa R, Kohno T, Totoki Y, et al. Expression and clinical significance of genes frequently mutated in small cell lung cancers defined by whole exome/RNA sequencing. Carcinogenesis 2015;36:616-21. [Crossref] [PubMed]
- Lu TP, Hsiao CK, Lai LC, et al. Identification of regulatory SNPs associated with genetic modifications in lung adenocarcinoma. BMC Res Notes 2015;8:92. [Crossref] [PubMed]
- Lu TP, Tsai MH, Lee JM, et al. Identification of a novel biomarker, SEMA5A, for non-small cell lung carcinoma in nonsmoking women. Cancer Epidemiol Biomarkers Prev 2010;19:2590-7. [Crossref] [PubMed]
- Sanchez-Palencia A, Gomez-Morales M, Gomez-Capilla JA, et al. Gene expression profiling reveals novel biomarkers in nonsmall cell lung cancer. Int J Cancer 2011;129:355-64. [Crossref] [PubMed]
- Quek K, Li J, Estecio M, et al. DNA methylation intratumor heterogeneity in localized lung adenocarcinomas. Oncotarget 2017;8:21994-2002. [Crossref] [PubMed]
- Fiori ME, Di Franco S, Villanova L, et al. Cancer-associated fibroblasts as abettors of tumor progression at the crossroads of EMT and therapy resistance. Mol Cancer 2019;18:70. [Crossref] [PubMed]
- Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene 2017;36:1461-73. [Crossref] [PubMed]
- Gelatti ACZ, Drilon A, Santini FC. Optimizing the sequencing of tyrosine kinase inhibitors (TKIs) in epidermal growth factor receptor (EGFR) mutation-positive non-small cell lung cancer (NSCLC). Lung Cancer 2019;137:113-22. [Crossref] [PubMed]
- Bikeye SN, Colin C, Marie Y, et al. ASPM-associated stem cell proliferation is involved in malignant progression of gliomas and constitutes an attractive therapeutic target. Cancer Cell Int 2010;10:1. [Crossref] [PubMed]
- Marie SK, Okamoto OK, Uno M, et al. Maternal embryonic leucine zipper kinase transcript abundance correlates with malignancy grade in human astrocytomas. Int J Cancer 2008;122:807-15. [Crossref] [PubMed]
- Tang J, Lu M, Cui Q, et al. Overexpression of ASPM, CDC20, and TTK Confer a Poorer Prognosis in Breast Cancer Identified by Gene Co-expression Network Analysis. Front Oncol 2019;9:310. [Crossref] [PubMed]
- Wang J, He Z, Duan R. Expression of ASPM in Lung Adenocarcinoma and Its Relationship with Development and Prognosis. Zhongguo Fei Ai Za Zhi 2020;23:29-35. [PubMed]
- Wang K, Chen R, Feng Z, et al. Identification of differentially expressed genes in non-small cell lung cancer. Aging 2019;11:11170-85. [Crossref] [PubMed]
- Pastushenko I, Brisebarre A, Sifrim A, et al. Identification of the tumour transition states occurring during EMT. Nature 2018;556:463-8. [Crossref] [PubMed]
- Li Z, Huang J, Shen S, et al. SIRT6 drives epithelial-to-mesenchymal transition and metastasis in non-small cell lung cancer via snail-dependent transrepression of KLF4. J Exp Clin Cancer Res 2018;37:323. [Crossref] [PubMed]
- Yang S, Liu Y, Li MY, et al. FOXP3 promotes tumor growth and metastasis by activating Wnt/beta-catenin signaling pathway and EMT in non-small cell lung cancer. Mol Cancer 2017;16:124. [Crossref] [PubMed]
(English Language Editor: R. Scott)


