
Genomic landscape of ground glass opacities (GGOs) in East Asians
Introduction
Lung adenocarcinoma (LUAD) is the most common histological subtype of cancer deaths worldwide (1). LUAD is the most common histologic subtype of lung cancer and is highly heterogeneity at histology and cellular level. Low-dose computed tomography (LDCT) guided screening has improved disease-related mortality by 20% (2). Surprisingly, the detection rate of small pulmonary nodules characterized by ground glass opacities (GGOs) has been greatly improved with the widespread implementation of CT (3). GGO was defined as hazy opacities with preserved bronchial and vascular margins in the lung parenchyma. Recently, an observation support the hypothesis that GGO proportion should be focused initial evaluation of histological subtypes in early stage LUADs (4). Smaller GGO proportion and nodules with lobulation or spiculation are determinants of poor prognosis histological subtypes in stage IA LUAD patients. GGOs are predominantly typical adenomatous hyperplasia (AAH), invasive adenocarcinoma (IAC), adenocarcinoma in situ (AIS), and minimally invasive adenocarcinoma (MIA) (5). It was reported that the 5-year survival rate was as high as 100% for resected AAH and AIS, while the 5-year survival rate for resected IAC was significantly reduced (6).
A comprehensive understanding of the cancer mutational events is a critical foundation for future diagnostics, prognostics, and targeted therapeutics.
The current diagnoses of AAH, IAC, AIS, and MIA are based on morphologic assessment. However, the disadvantage of this approach is that it cannot fully capture the potential biological characteristics of these pulmonary nodules. Epidermal Growth Factor Receptor (EGFR) mutations were frequently found in LUAD with GGO on high-dose CT (HRCT) (7). Previous multiple genomic studies have analyzed the diverse genomic landscape of GGOs (8-11) and identified core therapeutic targets, including EGFR (Epidermal Growth Factor Receptor) and ALK (Anaplastic Lymphoma Kinase). Despite some relevant observations, ancestral differences in GGO genomics have not been systematically elucidated due to the scarcity of resected specimens.
In order to deepen the understanding of the genomic landscape of GGOs, we sequenced the whole exomes (n=79) of 37 Chinese GGOs patients. Compared to previously published genomic landscapes of GGOs and LUADs, we depicted the comprehensive genomic landscape of GGOs and characterized complex ancestral differences, which may explain genomic evolution from preinvasive to invasive LUAD. In addition, we investigated the potential role of antitumor immune surveillance in the early development of LUAD through immunohistochemistry (IHC) analysis of antigen-specific CD8 T cells and programmed death ligand 1 (PD-L1) We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/jtd-21-82).
Methods
Patient recruitment and sample sequencing
Patients with GGOs were recruited from Tongji Hospital. Two specialized lung cancer pathologists were responsible for examination of the histologic subtypes, including MIA, AIS, and IAC. Due to the limited tissue samples obtained by surgical resection, AAH was not found in our study. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the institutional review board of Tongji Hospital and Huazhong University of Science and Technology, and all patients signed informed consent forms. Tumors and adjacent normal tissues were harvested by the pathologists following surgical resection. To ensure that the quality of the specimens met the requirements of next-generation sequencing (NGS), all tumor specimens were retrospectively reviewed by specialized lung cancer pathologists to determine the histological subtype.
Total deoxyribonucleic acid (DNA) was extracted from tissue samples collected from the patients. To ensure that more than 50% of the cells were tumor cells, the invasive tumor content was evaluated by pathologists. The DNA was extracted using Tissue Kit (69504, QIAGEN, Venlo, Netherlands). Targeted capture pulldown and exon-wide libraries were created from TruePrep DNA Library Prep Kit V2 for Illumina (#TD501, Vazyme, Nanjing, China) and native DNA using the xGen® Exome Research Panel (Integrated DNA Technologies, Inc., Skokie, Illinois, USA), and paired-end sequence data were generated using HiSeq 2500 platform (Illumina) with an average sequencing depth of 50× for normal tissues and 220× for tumor tissues. Reads were mapped to the human reference genome [National Center for Biotechnology Information (NCBI) build 37] with BWA, and polymerase chain reaction (PCR) duplicates were sorted and removed using sambamba (12).
Somatic variant identification
Single nucleotide variants (SNVs) were identified using MuTect (13). Somatic insertions and deletions (indels) were detected using Strelka2 (14). Somatic mutations were identified by statistical comparison, using the Fisher’s exact test as well as reference and non-reference reads in GGOs relative to those in corresponding normal tissues. A minimum of 20 reads covering the mutated region and five reads supporting the variant allele were required for somatic SNV/indel calling. In contrast, the sequencing depth needed to be ≥20×, and reads supporting the variant <5 at the same site in the normal control sample. Variants with MAF (minor allele frequency) >1% in the ExAC, gnomAD, and esp6500 databases were filtered out as common germline variants.
Driver gene identification and driver frequency comparison
The MutSigCV (15) algorithm was used to infer significantly mutated driver genes.
CNA (copy-number alteration) identification
Amplified and deleted regions of GGOs were identified with Genome Identification of Significant Targets in Cancer (GISTIC) (16) and the resulting copy number variations (CNVs) were used in further analysis. Somatic CNVs were analyzed using CNVkit (17). The genomic instability index (GII) was calculated as the percentage of the tumor genome.
Mutational signatures identification and clustering
The Nonnegative Matrix Factorization (NMF) (18) and MutationalPatterns (19) R packages were used to uncover the mutational processes of GGO patients. The resulting signature contribution proportions were further used to cluster samples using all known Catalogue Of Somatic Mutations In Cancer (COSMIC) signatures.
Histochemistry for PD-L1 and CD8 Analysis
The expression levels of PD-L1 and CD8 were independently scored by pathologists. For profiling analysis of PD-L1 expression, the proportion of PD-L1-positive tumor cells (TCs) or immune cells were quantified. Light microscopy (20) was used for the semiquantitative assessment for PD-L1 immunoreactivity. Membranous immunostaining was interpreted according to the proportion and intensity of positive tumor cells. Intensity was graded as 0 (negative), 1 (weak, <1%), 2 (moderate, 1–10%), or 3 (strong, 11–50% & >50%). The immunoreactive scores (IRS) were calculated from the sum of these values. An IRS value (≥3) was regarded as positive PD-L1 expression. The expression of CD8 on lymphocytes was recorded as the percentage of positive cells in nucleated cells in each nuclear compartment, and was defined as negative (≤10%) or positive (≥10%).
Statistical analysis
Statistical analysis was conducted using R packages.
Results
Patient samples and sequencing
We recruited 37 patients with GGOs and performed whole exome sequencing (WES) on 79 tumor tissues (Table S1). The average sequencing depth for WES was 229× (range, 120.4–383.2×), and 18,862 SNVs and 56 insertions/deletions (indels) were discovered in GGOs by Strelka2 (14) (https://cdn.amegroups.cn/static/public/jtd-21-82-1.xlsx). The mean tumor mutational burden (TMB) of East Asian-ancestry LUADs was 4.05 per megabase (median 2.04 per megabase) (21), which was much higher that of GGOs (mean 0.82 per megabase, median 0.66 per megabase) (Table S2).
Driver genes in GGOs
In this study, exome sequencing was used to describe the typical oncogene mutations and copy number changes during preneoplasia and preinvasive lung cancers. Through multi-region whole-exon sequencing, we focused on identifying driver genes, particularly those below the median of GGO patients. MutSigCV (15) identified significantly mutated genes among the 79 resected lung nodules including MIA (n=58), AIS (n=15), and IAC (n=6), along with 116 similarly-sequenced, previously reported pulmonary GGOs (9). Our study showed more frequent mutations in EGFR (22%), BRAF (v-RAF murine sarcoma viral oncogene homologue B1) (11%) and ERBB2 (Erb-B2 Receptor Tyrosine Kinase 2, also known as HER2) (8%), but less frequent mutations in driver genes KRAS (Kirsten rat sarcoma 2 viral oncogene homolog) (5%), MAP2K1 (mitogen-activated protein kinase 1, also known MEK1) (5%), NF1 (neurofibromatosis type 1) (5%), KMT2D (histone H3 lysine 4 methyltransferase) (4%), BCOR (BCL-6 corepressor) (3%) and MYH9 (Non-muscle myosin heavy chain IIA) (3%) (Figure 1A). Two indels were also discovered in GGOs, including EGFR and ERBB2. Unlike previous reports (9,10), our study did not identify two common tumor suppressor genes, STK11 (Serine Threonine Kinase 11) and CDKN2A (cyclin-dependent kinase inhibitor 2a), which are involved in chromosomal losses. Overall, we demonstrated that driver composition in patients with indeterminate pulmonary nodules (IPNs) (9) were different in our study. A major characteristic of GGOs was the low prevalence of driver genes, which was consistent with their lower TMB profile. When comparing driver genes across the LUADs (21) and IPNs (9), EGFR and KRAS were the most commonly mutated cancer genes. This is consistent with the notion that EGFR is a dominant driver gene of tumorigenesis (22).
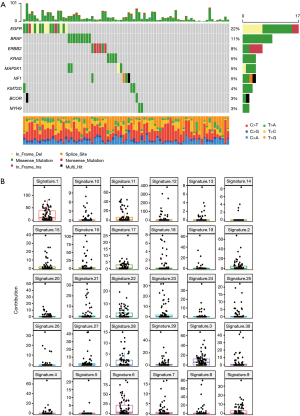
Mutational signatures of GGOs
Through mutational patterns buried within cancer genomes, the complexity and diversity of somatic cell mutational processes in human recessive carcinogenesis are being revealed. Mutational signatures are defined as different mutational processes that always generate different combinations of mutation types. The mutational signatures of GGOs were determined from COSMIC (available at https://cancer.sanger.ac.uk/cosmic/signatures);we discovered Signature 1, Signature 3, and Signature 6 in the patients with GGOs (Figure 1B).
Abnormalities in DNA maintenance may be related to mutational signatures. Signature 1 is thought to be related to the relative rate of increase of spontaneous deamination of 5-methylcytosine; Signature 3 is closely associated with somatic and germline BReast-CAncer susceptibility gene 1 (BRCA1) and BRCA2 mutations; and Signature 6 is characteristic of cancers with C > T at NpCpG mutations and defective DNA mismatch repair. In contrast, the mutation signatures of 210 patients with LUAD (21) were clustered into three signature groups including ‘aging’ (Signature 1), ‘APOBEC (apolipoprotein B mRNA editing catalytic polypeptide-like)’ (Signature 2), and ‘smoking’ (Signature 4). Our results further supported that Signature 1 was more prominent among early mutations, and the roles of Signature 3 and Signature 6 reveal the complexity of mutational processes behind cancer development, with potential significance for revealing cancer pathogenesis, prevention, and targeted therapy.
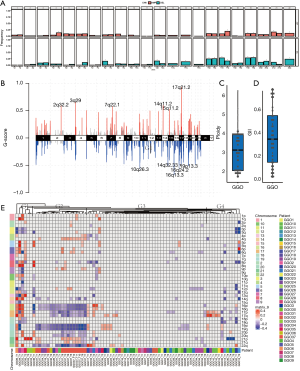
Ancestry differences of CNAs
CNVs reflect the instability of chromosomes. GISTIC was used to evaluate the chromosomal losses or gains of GGOs. The copy number gains or losses of chromosomal segments was defined as the chromosomal segments whose log2 ratio was less than –0.25 (loss) or greater than 0.25 (gain). Our study revealed that GGOs have large segments of amplification gain/loss. At the chromosomal level, the degree of arm-level CNV of GGOs patients was substantially lower than that of patients with LUAD (21) (Figure 2A). However, driver gene amplifications of 2q32.2, 3q29 (MUC4 [Mucin 4]), 7q22.1 (MUC12), 14q11.2, 15q11.2, and 17q21.2, and the deletions of 10q26.3, 14q32.33, 16p13.3, 16q24.2, and 19p13.3, were found in GGOs (Figure 2B). We further inferred CNAs for GGOs and calculated the ploidy and genomic instability index (GII). Compared to patients with LUAD (21), those with GGOs exhibited significant similarities in aneuploidy regardless of smoking history, and also showed lower GII, which was mainly related to fewer deletion events (Figure 2C,D). We extended our analysis to genome-wide CNA profiles of patients with GGOs. Interestingly, similar types of GGNs were clustered. As shown in Figure 2E, the cluster G1 had six MIA out of six members. Cluster G2 had 17 MIA out of 19 members and common regions of loss (16p-17q) and loss (19p-20q) were identified. However, cluster 4 had five AIS and two IAC out of 12 members. Overall, the results showed that GGOs had relatively fewer genomic changes and simpler genomic profiles than patients with LUAD.
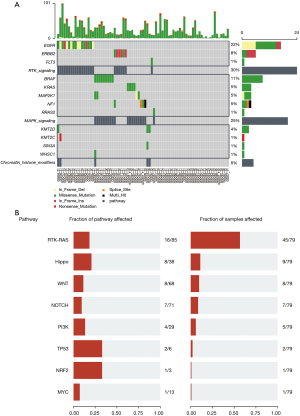
Therapeutic opportunities from ancestral differences
An overview of the genomic changes of GGOs may shed light on the potential differences in treatment. GGOs are characterized by recurrent aberrations in multiple key pathways and processes (Figure 3A). One core pathway with targetable genes, including EGFR (22%) and ERBB2 (8%), is the RTK/Ras/PI3K pathway. Combined with CNVs and mutations, GGOs showed significantly more somatic alterations including BRAF (11%), MAP2K1 (5%), and KRAS (5%), which were consistent with higher somatic alterations in LUADs. In contrast, EGFR mutations were consistently more common in GGOs with or without smoking status (21,22). Therefore, the RTK/Ras/PI3K pathway has a higher mutation frequency in GGOs (45/79). Examining a set of hallmark oncogenic pathways (23,24), GGOs also demonstrated a higher frequency of changes across the same pathways, including the Hippo (9/79) (25), Wnt (Wingless-related integration site) (8/79), Notch (7/79) (26), and PI3K (5/79) (27) pathways (Figure 3B).
Immune-relevant index analysis
In order to evaluate the role of T-cell immunomonitoring in 13 MIA, four AIS, and one IAC specimens, we performed IHC for PD-L1 and CD8 expression. As shown in Table S3, the positive expression of CD8 in 10 specimens was 56.0%, and seven of 13 MIA specimens had positive CD8 expression. The expression of PD-L1 in MIA (15.0%) was lower than that in AIS (25.0%). However, the positive expressions of PD-L1 and CD8 were lower than those in previous reports (8).
Discussion
The management of GGOs has gained increasing attention as these nodules may indicate lung cancer, most of which are adenocarcinomas. There were significant differences in size change during follow-up, frequency of coughing, smoking index, GGO size, location, distance to the pleura, presence of air bronchogram sign and positron emission tomography/computed tomography (PET/CT) appearance between the benign and malignant groups. Classifying GGOs as malignant or benign remains difficult. Therefore, as the proportion of benign GGOs was small in resected nodules, and the degree of malignancy was not low, a relatively active surgical strategy were recommend (28). And Relatively active surgical interventions could be considered for GGOs highly suspected of Malignancy.
Patients with solitary adenocarcinoma and multifocal ground-glass opacities (MGGOs) had a similar prognosis. Previous studies indicated that the presence of GGO components was found to be significantly associated with a favorable prognosis in clinical stage IA radiological invasive NSCLC (29,30). In addition, smoking history, type of MGGOs, histological types and predominant subtype of main tumor were independently significant prognostic factors for RFS in invasive stage I NSCLC (31,32). Recent report on the basic guidelines for the treatment of MGGO lesions is as follows. Firstly, a single-stage surgery was we performed to resect all lesions larger than 1 cm or solid-dominant lesions when tumors are located in the ipsilateral chest. For GGO-dominant lesions with slow growth or satellite lesions smaller than 1 cm, we perform strict surgical resection of the main tumor and easily accessible satellite lesions by limited resection, if the resection is expected to be larger than the sublober resection due to the central location or if multiple lesions are scattered throughout multiple lobes.
To evaluate pulmonary GGO and investigate the correlation between CT imaging features and LUAD driver genes or subtypes is helpful for diagnosis and clinical management. Pleural retraction, GGO proportion, and margin signature should be focused initial evaluation of histological subtypes in early stage LUADs (4). Different studies have different definition of GGO proportion and the following parameters have been used to calculate the GGO proportion: area ratio of tumor on mediastinal windows to that on lung windows, consolidation/tumor dimension ratio, GGO/tumor area ratio, the product of the dimension ratio of the tumor on mediastinal windows and GGO/tumor volume ratio (33) to calculate the tumor shadow disappearance rate (TDR) (34). However, many studies have reported that LUAD appears to differ significantly between individuals of European and Asian ancestry, with EGFR mutations present in only 7–10% of EUR LUADs but 40–60% of EAS LUADs (35,36). This further explains the uniqueness of GGO in the East Asian population.
Recently, there has been a significant increase in the detection of pulmonary nodules, many of which were MIA, AIS, preneoplasia atypical AAH, and IAC. Previous studies have revealed the complex genomic landscape of LUADs (21,37) and lung nodules (8-10,34) and identified many potential cancer driver genes, suggesting a driver role for these events in certain tumors. In our study, the characteristics of GGOs were identified by multi-region exome sequencing, which provided molecular evidence for the proposed model of early carcinogenesis of LUAD. Moreover, our study involved more patients with mutations in EGFR, BRAF, and ERBB2, further supporting the fact that EGFR was the most frequently mutated cancer gene in MIA, AIS, and IAC (9). Our founding also indicated that there was a high discrepancy of driver mutations in NSCLC patients with ground-glass nodules (GGNs) (4,38). EGFR mutation status including L858R mutation and 19 deletions, KRAS mutations or ALK rearrangements occurred frequently in patients with GGO(34). Somatic indels, such as EGFR and ERBB2, were also discovered in GGOs, which was consistent with previous reports (10). TP53 (tumor suppressor p53) was the most commonly mutated gene in numerous types of cancers including LUADs (21), while TP53 (1%) was a low-frequency mutation in GGOs (Figure S1). Contrary to previous research on pulmonary nodules (8,9), common tumor suppressor genes, including STK11 and CDKN2A, were not found. However, the genomic landscape of LUAD (21) showed common driver genes such as KRAS, NF1, and BRAF. The carcinogenesis of lung cancer may be the result of an accumulation of mutations in a branched evolutionary pattern, like a growing tree (39). Our results indicated that there are similar genetic profiles between LUAD and GGOs, which further highlights the genomic evolution from preneoplasia lesions to invasive LUAD.
In order to further reveal the mutational patterns buried within cancer genomes, GISTIC identified significant aberrations in Signature 1, Signature 3, and Signature 6 in the patients with GGOs. Compared with the three signatures extracted from LUADs (21,35), Signature 1 was more prominent among GGO patients, and was associated with cigarette smoking. However, no somatic mutations in DNA repair genes, including BRCA1 and BRCA2, could explain the occurrence of aberrations in Signature 3. When compared with the three signatures extracted from LUAD, Signature 2 was unapparent in GGO patients, suggesting that the APOBEC signatures were enriched in late mutations (40). This observation indicated that these three signatures were significantly associated with GGOs, and showed diversity in different stages of LUAD. Furthermore, compared with LUADs, GGOs seem to more often carry a dominant proportion of ‘Signature 1’. Genome doubling and ongoing dynamic chromosomal instability indicated intratumor heterogeneity in GGOs. Compared with LUADs (21), GGOs exhibited significantly lower levers of arm-level CNV (Figure 2A), and lower focal CNVs (Figure 2B). Our results revealed that GGOs had fewer genomic alterations and simpler genomic profiles than LUADs.
Previous studies have shown that that somatic signaling pathways occurred in cancer at varying frequencies and in varying combinations across different organs and tissues. GGOs had repeated aberrations in some key pathways, and some targeted driver mutations, including those in EGFR, KRAS, and BRAF, were almost clonal and early, explaining the robust and consistent responses that are often seen across multiple aspects of the disease with regard to these targeted alterations. Moreover, signaling pathway studies have revealed that the most commonly altered processes of GGOs are related to the RTK/Ras/PI3K (mTOR) pathways, and EGFR alterations were the dominant genetic changes across all targetable somatic changes
Acknowledgments
The authors would like to thank Shanghai Tongshu Biotechnology Co., Ltd. for their technical support.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/jtd-21-82
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jtd-21-82
Disclosure: All chart data are from WES data except Tables S1 and S3 for statistical analysis.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-21-82). All authors report funding from Hubei Natural Science and Technology Fund Youth Fund (No. CFB295). The authors have no other conflict of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the institutional review board of Tongji Hospital and Huazhong University of Science and Technology, and the reference number for approval was TJ-C20190602. Written consent was obtained from the patients for all forms of personally identifiable data, including biomedical, clinical, and biometric data.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- National Lung Screening Trial Research Team. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011;365:395-409. [Crossref] [PubMed]
- Bongiolatti S, Corzani R, Borgianni S, et al. Long-term results after surgical treatment of the dominant lung adenocarcinoma associated with ground-glass opacities. J Thorac Dis 2018;10:4838-48. [Crossref] [PubMed]
- Miao Y, Zhang J, Zou J, et al. Correlation in histological subtypes with high resolution computed tomography signatures of early stage lung adenocarcinoma. Transl Lung Cancer Res 2017;6:14-22. [Crossref] [PubMed]
- From the American Association of Neurological Surgeons (AANS), American Society of Neuroradiology (ASNR), Cardiovascular and Interventional Radiology Society of Europe (CIRSE), et al. Multisociety Consensus Quality Improvement Revised Consensus Statement for Endovascular Therapy of Acute Ischemic Stroke. Int J Stroke 2018;13:612-32. [PubMed]
- Travis WD, Brambilla E, Noguchi M, et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol 2011;6:244-85. [Crossref] [PubMed]
- Yoshida Y, Kokubu A, Suzuki K, et al. Molecular markers and changes of computed tomography appearance in lung adenocarcinoma with ground-glass opacity. Jpn J Clin Oncol 2007;37:907-12. [Crossref] [PubMed]
- Zhang C, Zhang J, Xu FP, et al. Genomic Landscape and Immune Microenvironment Features of Preinvasive and Early Invasive Lung Adenocarcinoma. J Thorac Oncol 2019;14:1912-23. [Crossref] [PubMed]
- Hu X, Fujimoto J, Ying L, et al. Multi-region exome sequencing reveals genomic evolution from preneoplasia to lung adenocarcinoma. Nat Commun 2019;10:2978. [Crossref] [PubMed]
- Ren Y, Huang S, Dai C, et al. Germline Predisposition and Copy Number Alteration in Pre-stage Lung Adenocarcinomas Presenting as Ground-Glass Nodules. Front Oncol 2019;9:288. [Crossref] [PubMed]
- Kobayashi Y, Mitsudomi T, Sakao Y, et al. Genetic features of pulmonary adenocarcinoma presenting with ground-glass nodules: the differences between nodules with and without growth. Ann Oncol 2015;26:156-61. [Crossref] [PubMed]
- Tarasov A, Vilella AJ, Cuppen E, et al. Sambamba: fast processing of NGS alignment formats. Bioinformatics 2015;31:2032-4. [Crossref] [PubMed]
- Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213-9. [Crossref] [PubMed]
- Kim S, Scheffler K, Halpern AL, et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat Methods 2018;15:591-94. [Crossref] [PubMed]
- Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013;499:214-8. [Crossref] [PubMed]
- Mermel CH, Schumacher SE, Hill B, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 2011;12:R41. [Crossref] [PubMed]
- Talevich E, Shain AH, Botton T, et al. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS Comput Biol 2016;12:e1004873 [Crossref] [PubMed]
- Gaujoux R, Seoighe C. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics 2010;11:367. [Crossref] [PubMed]
- Blokzijl F, Janssen R, van Boxtel R, et al. MutationalPatterns: comprehensive genome-wide analysis of mutational processes. Genome Med 2018;10:33. [Crossref] [PubMed]
- Noh BJ, Kwak JY, Eom DW. Immune classification for the PD-L1 expression and tumour-infiltrating lymphocytes in colorectal adenocarcinoma. BMC Cancer 2020;20:58. [Crossref] [PubMed]
- Chen J, Yang H, Teo A, et al. Genomic landscape of lung adenocarcinoma in East Asians. Nat Genet 2020;52:177-86. [Crossref] [PubMed]
- Nahar R, Zhai W, Zhang T, et al. Elucidating the genomic architecture of Asian EGFR-mutant lung adenocarcinoma through multi-region exome sequencing. Nat Commun 2018;9:216. [Crossref] [PubMed]
- Sanchez-Vega F, Mina M, Armenia J, et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018;173:321-37.e10. [Crossref] [PubMed]
- Martínez-Jiménez F, Muiños F, Sentís I, et al. A compendium of mutational cancer driver genes. Nat Rev Cancer 2020;20:555-72. [Crossref] [PubMed]
- Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer 2013;13:246-57. [Crossref] [PubMed]
- Takebe N, Miele L, Harris PJ, et al. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat Rev Clin Oncol 2015;12:445-64. [Crossref] [PubMed]
- Vasan N, Toska E, Scaltriti M. Overview of the relevance of PI3K pathway in HR-positive breast cancer. Ann Oncol 2019;30:x3-x11. [Crossref]
- Qin Y, Xu Y, Ma D, et al. Clinical characteristics of resected solitary ground-glass opacities: Comparison between benign and malignant nodules. Thorac Cancer 2020;11:2767-74. [Crossref] [PubMed]
- Hattori A, Matsunaga T, Hayashi T, et al. Prognostic Impact of the Findings on Thin-Section Computed Tomography in Patients with Subcentimeter Non-Small Cell Lung Cancer. J Thorac Oncol 2017;12:954-62. [Crossref] [PubMed]
- Hattori A, Matsunaga T, Takamochi K, et al. Importance of Ground Glass Opacity Component in Clinical Stage IA Radiologic Invasive Lung Cancer. Ann Thorac Surg 2017;104:313-20. [Crossref] [PubMed]
- Shimada Y, Maehara S, Kudo Y, et al. Profiles of Lung Adenocarcinoma With Multiple Ground-Glass Opacities and the Fate of Residual Lesions. Ann Thorac Surg 2020;109:1722-30. [Crossref] [PubMed]
- Fu F, Zhang Y, Wen Z, et al. Distinct Prognostic Factors in Patients with Stage I Non-Small Cell Lung Cancer with Radiologic Part-Solid or Solid Lesions. J Thorac Oncol 2019;14:2133-42. [Crossref] [PubMed]
- Usuda K, Sagawa M, Motono N, et al. Relationships between EGFR mutation status of lung cancer and preoperative factors - are they predictive. Asian Pac J Cancer Prev 2014;15:657-62. [Crossref] [PubMed]
- Gao JW, Rizzo S, Ma LH, et al. Pulmonary ground-glass opacity: computed tomography features, histopathology and molecular pathology. Transl Lung Cancer Res 2017;6:68-75. [Crossref] [PubMed]
- Wu K, Zhang X, Li F, et al. Frequent alterations in cytoskeleton remodelling genes in primary and metastatic lung adenocarcinomas. Nat Commun 2015;6:10131. [Crossref] [PubMed]
- Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst 2005;97:339-46. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Wu C, Zhao C, Yang Y, et al. High Discrepancy of Driver Mutations in Patients with NSCLC and Synchronous Multiple Lung Ground-Glass Nodules. J Thorac Oncol 2015;10:778-83. [Crossref] [PubMed]
- Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med 2017;376:2109-21. [Crossref] [PubMed]
- de Bruin EC, McGranahan N, Mitter R, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 2014;346:251-6. [Crossref] [PubMed]
(English Language Editor: A. Kassem)


