
Knockdown of lncRNA TUC338 inhibits esophageal cancer cells migration and invasion
Introduction
Esophageal disease is quite possibly the most well-known harmful tumors of the upper gastrointestinal tract with more than 300,000 new patients every year, posing a genuine danger to human wellbeing (1). Esophageal Squamous Cell Carcinoma (ESCC) is the primary histological sort, accounting approximately 80% of esophageal malignant growth. With the advancements in early tumor diagnosis and treatment technology, the 5-year survival rate of esophageal cancer stays at 15–25%. This is on account of the combined effect of nearby intrusion and far off metastasis. Numerous patients with esophageal disease have already appeared metastasis at the first diagnosis (2,3). Complications caused by tumor metastasis are the main cause of cancer-related deaths. In recent years, numerous examinations have discovered that quality articulation, cytokine discharge, epithelial-mesenchymal transition (EMT), and changes in the tumor ambient are closely related to tumor metastasis, but this complex process can only be partially clarified in specific tumor types (4). Therefore, elucidating the molecular regulatory mechanisms and important targets of tumor metastasis will provide new opportunities for tumor treatment. An enormous number of studies have shown that EMT assumes a significant part in tumor movement, metastasis, and medication obstruction. EMT is an interaction by which epithelial cells lose apex-basal polarity and cell-cell attachment, and progress to forceful mesenchymal cells (5). EMT is associated with many organic and neurotic cycles, including embryonic turn of events, wound mending, malignant growth cell metastasis, and medication opposition. During the process of EMT, the articulation levels of epithelial qualities, (for example, E-cadherin) are reduced, and metaplastic gene (N-cadherin, vimentin) expression levels increase. Much of the time, the deficiency of E-cadherin is an indication of EMT (6,7). The transcription factor Slug play vital roles in the development of motile and invasive manner of cancer cells via EMT progression (7). Changes in quality articulation during EMT lead to numerous phenotypic changes, for example, changes in cell morphology, loss of grip, and acquisition of stem cell like attributes (8).
Long non-coding RNAs (lncRNAs) are sorts of records with lengths more prominent than 200 nt, which can’t be converted into proteins in cells (9). A large number of studies have revealed that lncRNA assumes a significant part in different cell cycles like cell separation, multiplication, migration, and invasion (10). It is worth noting that when lncRNA is abnormally expressed in cells, it can lead to the occurrence of many diseases, including diabetes, cardiovascular disease, and tumors (9,11). Studies have shown that lncRNA assumes a significant part in the event, advancement, and guess of tumors. In esophageal cancer, lncRNAs H19, DNM3OS, and SNHG7 perform physiological regulatory functions by regulating intermediate miRNAs, including epigenetic modification and transcription regulation, amongst others (12-14). These discoveries demonstrate that lncRNAs might be utilized as biomarkers for metastasis observing and might be utilized as restorative focuses to restrain metastasis.
LncRNA TUC338 has recently been identified as part of a class of cancer-promoting genes, however, its job in esophageal cancer is indistinct. This investigation preliminarily analyzed the declaration of TUC338 in esophageal malignant growth tissues and its relationship with the facility obsessive qualities of patients, also explored the effect of TUC338 on the expansion and migration of esophageal malignant growth cells and related molecular mechanisms. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/jtd-21-563).
Methods
Clinical sample collection
The 100 esophageal cancer tissue samples used in this experiment were surveyed and approved by the Ethics Committee of Jiangsu Cancer Hospital. The collection for all tissue samples was carried out with the patient’s informed consent, and the informed consent and tissue sample consent were signed in accordance with the process. Samples were collected between January 2014 and August 2019. The excised cancer tissue was taken from the operating room, washed twice with pre-cooled phosphate buffered saline containing double antibodies, and use blotting paper to dip water on the tissue. Carefully separate tumor tissue and adjacent tissues on ice, and a portion was transferred to a 1.5-mL centrifuge tube with RNA protection solution. Samples were put away in fluid nitrogen for later use in the future.
All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Jiangsu Cancer Hospital (No.:2013-840) and informed consent was taken from all the patients.
Cell cultures
Human esophageal malignant growth cell lines EC9706, KYSEl80, KYSE450, KYSE150, and EC109 were bought from the Shanghai Cell Bank of the Chinese Academy of Sciences. The ordinary human esophageal epithelial cell line Het-1A was bought from American type culture collection (ATCC). Esophageal malignant growth cell were refined with 1640 medium, which contained 10% fetuses bovine serum and 1% twofold anti-microbial (streptomycin-penicillin), and incubated in a 37 °C cell incubator (5% CO2). The cells were passaged in 2–3 days, and the way of life were extended by the proportion of 1:2.
RNA extraction and RT-qPCR
Complete RNA was removed from 50 mg of tissues and cells preserved in liquid nitrogen by adding 1 mL Trizol using an absolute RNA extraction unit, as per the maker’s guidelines. Invert record was performed utilizing a converse record pack to acquire cDNA. The SYBR Green color strategy was utilized for the RT-PCR response. The △△Ct technique was utilized to ascertain the declaration of TUC338, and 18S rRNA was utilized as an interior control (15). The preliminary sequences were: 5'-GGTGAGAGGGGATGTTCAGT-3' (onward) and 5'-TGGGTGAAATGAGGTTG-3' (backward) for TUC338; 5'-CGCTTCCTTACCTGGTTGAT-3' (onward) and 5'-GAGCGACCAAAGGAACGATA-3' (backward) to 18S rRNA.
Cell transfection
The shRNA targeting TUC338 (shRNA: 5’-CCACAGGACAGGUACAGCATT-3’) and the negative control oligonucleotide were synthesized by Shanghai Gema Gene Company. The shRNA sequences and negative sequence were used according to the references (16). For RNA interference, cells were cultivated in a 6-well plate at a thickness of 3×105/all things considered, and afterward transfected with Lipofectamine 2000-encapsulated shRNA or negative regulation. At 48 h after transfection, the cells were collected to extract RNA, and qRT-PCR was utilized to distinguish the impedance productivity.
CCK-8 to detect cell viability
EC109 cells were gathered 48 h after transfection, processed with trypsin, and seeded into 96-well plates at a thickness of 2×103/well. In the wake of refined for 24, 48, 72, and 96 h, 10 µL of CCK-8 stock arrangement was added to each well, at that point cells were brooded at 37 °C with 5% CO2 to 3 h. OD values were then detected at a wavelength of 490 nm.
Plate clone formation experiment
EC109 cells was collected 48 h after transfection, digested with trypsin, and immunized into 6-well plates at a thickness of 600/well. After incubating for 14 days in a 37 °C, 5% CO2 incubator, cells were stabled with 1% paraformaldehyde to 10 minutes. Cells were washed with PBS 3 times, treated with 0.1% crystal violet for 15 minutes, and put under the magnifying lens for tallying and investigation.
Transwell experiments
Migration experiment
EC109 cells were collected 48 h after transfection. After digestion with trypsin, the cells were diluted with medium containing 1% fetal bovine serum, then inoculated in the upper office of the Transwell at a thickness of 2×104/well. Then, 1 mL of complete medium was added to cells, which were then stored in a 37 °C, 5% CO2 hatchery for 24 h. Cells were stable and spatter, at that point the cells inside the Transwell was gently wiped with a cotton swab. Images were captured and calculations were made under a microscope.
Invasion experiment
After the transfection EC109 cells were gathered for 48 h. After digestion with trypsin, the cells were diluted with 1% medium containing fetal ox-like serum to change the cell thickness. Next, 100 µL Matrigel was added to the upper office of the Transwell, which was then positioned in a 4 °C refrigerator for 30 minutes. EC109 cells were then vaccinated in the upper officer at a density of 2×104/well, what’s more, 1 mL of complete medium was added to the Transwell chamber and put in a 37 °C, 5% CO2 hatchery for 24 h. Cells were stabled and blemish, at that point the cells inside the Transwell chamber were delicately cleaned with a cotton swab. Pictures were caught and estimations were made under a magnifying lens.
Western blot detection
EC109 cells 48 h after transfection were collected, lysed with protein lysis buffer on ice, and protein fixation was resolved utilizing the BCA protein measurement strategy. An example measure of 50 µg was added at each well. A 12% polyacrylamide gel was utilized for protein electrophoresis, then electrophoresis voltage was applied to 90 and 120 V steady pressing factor. After consummation, wet exchange to the PVDF film was performed, and the layer was brooded in PBS. Membranes were then incubated in 5% cream milk powder at room temperature for 2 h, at that point hatched with the essential antibody overnight at 4 °C. GAPDH was the internal reference protein. A 1:2,000 peroxidase-labeled secondary antibody was then added, and the membrane was hatched at room temperature for 2 h. The layer was put in a gel imager to catch pictures after color development, and image analysis was performed.
Statistical analysis
The SPSS20.0 measurable programming was utilized to examine the information, and the information were communicated as mean ± standard deviation (SD). Correlations between bunches were performed utilizing the t test or investigation of difference, and P<0.05 was considered measurably huge.
Results
The relationship between the high expression of TUC338 in esophageal cancer and the clinic pathological characteristics of patients Studies have shown that TUC338 is profoundly communicated in tongue cancer, and promotes cell proliferation while inhibiting apoptosis. Therefore, we firstly collected 100 instances of esophageal malignant growth tissues and comparing contiguous tissues, and detected the expression of TUC338 by fluorescence quantitative PCR. The results showed that the articulation level of TUC338 in esophageal cancer tissue was altogether higher than that of adjoining tissues (Figure 1A). Based on the above examination results, and according to the expression level of TUC338, we divided the esophageal cancer tissues into the TUC338 high articulation gathering and TUC338 low articulation bunch, and analyzed the correlations with the clinic pathological characteristics of the patients. Result states that TUC338 high articulation gathering was correlated with tumor size (Table 1). Using Kaplan-Meier survival analysis to further inspect the relationship in the middle of TUC338 articulation and patient endurance, the outcomes showed that patients with high TUC338 articulation had a more limited endurance period (Figure 1B). The above results show that the articulation level of TUC338 can be utilized as a marker for the conclusion and forecast of esophageal malignancy.

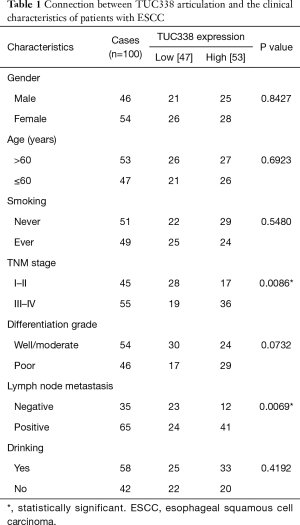
Full table
Knockdown of TUC338 can altogether restrain the expansion and clone arrangement of esophageal malignant cells
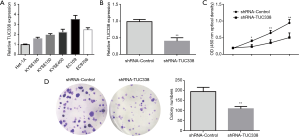
The above experimental results show that TUC338 is distinctly indicated in esophageal malignant tissues. to develop this study, effect of TUC338 on the function of esophageal malignant cells, we first performed fluorescence quantitative PCR to discover the uttering levels of TUC338 under common esophageal malignancy cell lines. The effect intimated: Compared with other cancer cell line (EC9706, KYSEl80, KYSE450 and KYSE150), TUC338 was highly expressed in EC109 cells (Figure 2A). Therefore, the constructed shRNA was transfected into EC109 cells, & the reaction reveals that the transfection of shRNA could remarkably reduce the articulation level of TUC338 in group (Figure 2B). The product of CCK-8 experiments showed that compared with shRNA-NC, knocking down TUC338 essentially repressed the expansion of EC109 esophageal malignant cells (Figure 2C). Additionally, the plate clone formation experiment showed that contrasted along the shRNA-NC category, knocking down TUC338 significantly inhibited the number of colony formations (Figure 2D). The above results show that knocking down TUC338 can fundamentally hinder the multiplication and clone arrangement capacity of esophageal malignancy cells.
Knockdown of TUC338 can significantly inhibit the relocation and intrusion of esophageal malignancy cells
The relocation and intrusion abilities of tumor cells are the main features of malignant tumors, and it is also the prerequisite for tumor cell metastasis to distant sites. Therefore, we used a Transwell assay to further test the effect of knocking down TUC338 on the intrusion & migration of esophageal malignant cells. First, the transfected EC109 cells were inoculated in the top chamber of the Transwell, and then fixed, stained, and analyzed 24 hours later. The results showed that knocking down TUC338 altogether hindered the development of EC109 esophageal malignant growth cells compared with the blank control group (shRNA-NC) (Figure 3A). To assess the impact of TUC338 knockdown on cell invasion ability, Matrigel was added to the upper Transwell chamber and the transfected EC109 cells were inoculated in the upper chamber for 24 hours, followed by fixation, staining, and analysis. The results showed that compared with shRNA-NC, knocking down TUC338 notably reluctant the migration capability of EC109 esophageal malignant cells (Figure 3B).

Knockdown of TUC338 can significantly inhibit the expression of EMT-related molecules in esophageal cancer cells
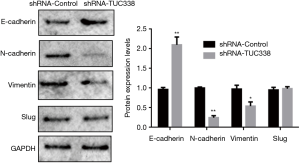
EMT is a significant natural cycle by which dangerous tumor cells got from epithelial cells gain relocation and attack capacities. Its principle qualities incorporate the diminished articulation of cell grip particles, (for example, E-cadherin) and increased vimentin. To additional examination the impact of TUC338 knockdown on the molecular mechanisms of esophageal malignancy cells, we performed western smudge discovery of the expression of cell invasion-related molecules. The results showed that in contrast with the blank control, knocking down TUC338 notably inhibited the expression of vimentin and N-cadherin in EC109 esophageal malignant cells, while the assertion of E-cadherin increased in TUC338 knockdown cells. There was no significant effect on the expression of the transcription factor Slug (Figure 4). This indicates that knocking down TUC338 can significantly inhibit EMT processes in esophageal cancer.
Discussion
ESCC has one of the highest mortality rates in the world. Although current research has provided a certain understanding of the pathogenesis and treatment of ESCC, as well as a preliminary understanding of the expression levels and effects of TUC338 in liver cancer, tongue squamous cell carcinoma, and lung cancer, the jobs and administrative mechanisms of lncRNA TUC338 in ESCC prevail unclear. This investigation firstly found that TUC338 is exceptionally communicated in esophageal malignant tissues and can be used as an analytic and prognostic marker of esophageal malignancy. Furthermore, knockdown of TUC338 fundamentally restrained the multiplication and attack of esophageal malignant cells.
Research has shown that non-coding RNAs have different expression profiles in different tumors. Recent studies have shown that TUC338 can advance the event and improvement of tumors as an oncogene. For example, TUC338 is thoroughly demonstrate in liver malignant cell lines and tissues, and promotes cell proliferation by regulating the cell cycle (17). In addition, TUC338 is up-regulated in patients with early bladder cancer, and the level of TUC338 in plasma is significantly reduced after surgical resection, which suggests that TUC338 could be utilize as a fluid detection marker for the early conclusion and postoperative identification of bladder cancer (18). TUC338 is exceptionally communicated, more promote prostate cancer by down-regulating miR-466 (15). TUC338 also plays a significant part in the event and movement of non-small cell lung cancer (NSCLC), and can be used as a potential independent prognostic biomarker for NSCLC patients (19). The expression of TUC338 in esophageal cancer has remained unclear until now. The results of this study show that TUC338 is extensively communicated in clinic pathological, and the esophageal Cancer tissues characteristics of patients such as TNM stage are significantly related to metastasis. The endurance season of patients with high TUC338 articulation is altogether lower than that of patients with low articulation. These results suggest that TUC338 can be utilized as a marker for early analysis and anticipation of esophageal cancer. Since TUC338 can be detected in the peripheral blood of patients, we will continue to collect peripheral blood samples with different case characteristics in subsequent experiments to evaluate the feasibility and specificity of TUC338 as a liquid biopsy marker to detect the progression of esophageal cancer.
The malignant and disordered proliferation of cells is a main feature of tumors, and inhibition of proliferation has also become the main research direction and goal for the treatment of tumors. To additionally investigate the particular elements of TUC338 into esophageal cancer, we knocked down TUC338 in cells to assess its role in cell functions. The results show that knocking down TUC338 can essentially repress the expansion and settlement development capacity of cells. This is steady with the aftereffects of other past examinations. In tongue squamous cell carcinoma, silencing TUC338 significantly restrained cell development and expanded cell apoptosis (20). In liver disease cells, TUC338 can manage cell expansion and change cell development in a way like record factors. For example, TUC338 can bind to Plasminogen Activator Inhibitor-1 RNA Binding Protein (PAI-RBP1) to regulate its expression (21). In addition, TUC338 can also play a role in the drug resistance of tumor tissues. For example, TUC338 participates in the function of sorafenib-sensitized liver cancer cells by targeting RASAL1 (22). Therefore, we speculate that TUC338 can be combined with other chemotherapeutics to reverse the drug resistance of tumor cells.
EMT is the cycle by which epithelial cells acquire mesenchymal characteristics. In cancer, EMT is related to tumor occurrence, invasion, metastasis, and resistance to treatment. Many patients with esophageal cancer have metastasis when they are first diagnosed. Complications caused by tumor metastasis are the fundamental driver of death in patients with esophageal disease. Past examinations have shown that TUC338 inhibits the migration and invasion of cervical cancer cells by targeting the TIMP1 gene (23). In lung cancer, TUC338 inhibits cell invasion and migration by regulating the MAPK signaling pathway (16). The results of this study show that knocking down TUC338 can altogether restrain the relocation and attack of esophageal cancer cells. At the simultaneously, western blot detection showed that TUC338 could articulation of vimentin and N-cadherin, increment the outflow of E-cadherin, and repress the EMT interaction.
In outline, this examination found that TUC338 is profoundly communicated in esophageal malignant growth tissues, and is correlated with patients’ clinic pathological characteristics (TNM stage and lymphatic metastasis). Cell function experiments proved that knocking down TUC338 could significantly inhibit cell proliferation and colony formation. At the same time, knocking down TUC338 increased the articulation of E-cadherin by suppressing the articulation of vimentin and N-cadherin, thereby inhibiting tumor cell migration and invasion.
Acknowledgments
Funding: This work was supported by the scientific research project of Jiangsu Cancer Hospital (Grant No. ZM202012) and Natural Science Foundation of Jiangsu Province, China (Grant No. BK20201496)..
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/jtd-21-563
Data Sharing Statement: Available at http://dx.doi.org/10.21037/jtd-21-563
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/jtd-21-563). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Jiangsu Cancer Hospital (No.:2013-840) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
Reference
- Alsop BR, Sharma P. Esophageal Cancer. Gastroenterol Clin North Am 2016;45:399-412. [Crossref] [PubMed]
- Kato H, Nakajima M. Treatments for esophageal cancer: a review. Gen Thorac Cardiovasc Surg 2013;61:330-5. [Crossref] [PubMed]
- Yu Z, Li XM, Huai M, et al. Clinic features and prognostic analysis for T1 esophagus cancer. Zhonghua Zhong Liu Za Zhi 2018;40:268-73. [PubMed]
- Abbasi BA, Iqbal J, Ahmad R, et al. Potential phytochemicals in the prevention and treatment of esophagus cancer: A green therapeutic approach. Pharmacol Rep 2019;71:644-52. [Crossref] [PubMed]
- Liu J, Li C, Zhang L, et al. Association of tumour-associated macrophages with cancer cell EMT, invasion, and metastasis of Kazakh oesophageal squamous cell cancer. Diagn Pathol 2019;14:55. [Crossref] [PubMed]
- De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer 2013;13:97-110. [Crossref] [PubMed]
- Goossens S, Vandamme N, Van Vlierberghe P, et al. EMT transcription factors in cancer development re-evaluated: Beyond EMT and MET. Biochim Biophys Acta Rev Cancer 2017;1868:584-91. [Crossref] [PubMed]
- Pastushenko I, Blanpain C. EMT Transition States during Tumor Progression and Metastasis. Trends Cell Biol 2019;29:212-26. [Crossref] [PubMed]
- Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 2017;36:5661-7. [Crossref] [PubMed]
- Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res 2017;77:3965-81. [Crossref] [PubMed]
- Kumar MM, Goyal R. LncRNA as a Therapeutic Target for Angiogenesis. Curr Top Med Chem 2017;17:1750-7. [Crossref] [PubMed]
- Chen MJ, Deng J, Chen C, et al. LncRNA H19 promotes epithelial mesenchymal transition and metastasis of esophageal cancer via STAT3/EZH2 axis. Int J Biochem Cell Biol 2019;113:27-36. [Crossref] [PubMed]
- Zhang H, Hua Y, Jiang Z, et al. Cancer-associated Fibroblast-promoted LncRNA DNM3OS Confers Radioresistance by Regulating DNA Damage Response in Esophageal Squamous Cell Carcinoma. Clin Cancer Res 2019;25:1989-2000. [Crossref] [PubMed]
- Xu LJ, Yu XJ, Wei B, et al. LncRNA SNHG7 promotes the proliferation of esophageal cancer cells and inhibits its apoptosis. Eur Rev Med Pharmacol Sci 2018;22:2653-61. [PubMed]
- Li G, Zhang Y, Mao J, et al. LncRNA TUC338 is overexpressed in prostate carcinoma and downregulates miR-466. Gene 2019;707:224-30. [Crossref] [PubMed]
- Zhang YX, Yuan J, Gao ZM, et al. LncRNA TUC338 promotes invasion of lung cancer by activating MAPK pathway. Eur Rev Med Pharmacol Sci 2018;22:443-9. [PubMed]
- Braconi C, Valeri N, Kogure T, et al. Expression and functional role of a transcribed noncoding RNA with an ultraconserved element in hepatocellular carcinoma. Proc Natl Acad Sci U S A 2011;108:786-91. [Crossref] [PubMed]
- Li G, Zhang Y, Mao J, et al. lncRNA TUC338 is a potential diagnostic biomarker for bladder cancer. J Cell Biochem 2019;120:18014-9. [Crossref] [PubMed]
- Tian Y, Feng Y. Up-regulation of long noncoding RNA uc.338 predicts poor survival in non-small cell lung cancer. Cancer Biomark 2018;22:781-5. [Crossref] [PubMed]
- Ouyang KX, Zou R, Liang J, et al. TUC338 Overexpression Leads to Enhanced Proliferation and Reduced Apoptosis in Tongue Squamous Cell Carcinoma Cells In Vitro. J Oral Maxillofac Surg 2017;75:423-8. [Crossref] [PubMed]
- Wen HJ, Walsh MP, Yan IK, et al. Functional Modulation of Gene Expression by Ultraconserved Long Non-coding RNA TUC338 during Growth of Human Hepatocellular Carcinoma. iScience 2018;2:210-20. [Crossref] [PubMed]
- Jin W, Chen L, Cai X, et al. Long non-coding RNA TUC338 is functionally involved in sorafenib-sensitized hepatocarcinoma cells by targeting RASAL1. Oncol Rep 2017;37:273-80. [Crossref] [PubMed]
- Li Q, Shen F, Wang C. TUC338 promotes cell migration and invasion by targeting TIMP1 in cervical cancer. Oncol Lett 2017;13:4526-32. [Crossref] [PubMed]
(English Language Editor: C. Betlazar-Maseh)


