
Genetic variants in Chinese patients with sporadic Stanford type A aortic dissection
Introduction
Aortic dissection (AD) is a catastrophic cardiovascular condition that involves separation of the layers of the aortic wall. Stanford type A AD (STAAD) involves the ascending aorta, and it is further divided into DeBakey type I or type II based on the extent of the dissection (1,2). Patients with STAAD have a stronger genetic component than patients with Stanford type B AD or abdominal aortic aneurysms, which are more associated with lifestyle-linked risk factors, such as hypertension, atherosclerosis, age and sex (3,4).
Genetic disorders are strongly associated with aortic disease, and genetic disorder-induced aortic wall weakness may cause aortic aneurysms and dissection. Approximately 11–19% of patients with AD have first-degree relatives who are diagnosed with aortic aneurysm or dissection, and 80% of patients show a sporadic form of AD (5,6). Patients with aortic aneurysms and dissections are further classified as nonsyndromic or syndromic according to whether the abnormalities are limited to the cardiovascular system. Hereditary diseases of connective tissues are often associated with syndromic AD, and most of these patients develop an autosomal dominant disorder caused by mutations in certain genes, such as fibrillin-1 [(FBN1); Marfan syndrome (MFS)], fibrillin-2 [(FBN2); Beals syndrome], collagen type III alpha 1 chain [(COL3A1); Ehlers-Danlos syndrome (EDS), vascular type] and transforming growth factor beta receptor [(TGFBR); Loeys-Dietz syndrome (LDS)], whereas mutations located in acetyl coA acetyltransferase 2 (ACAT2), myosin heavy chain 11 (MYH11) and SMAD2 seem to affect patients with a family history and exhibit a nonsyndromic form (7). However, the genetic risk factors for patients with sporadic STAAD are not clear.
Next-generation sequencing (NGS) technology is widely used for clinical testing in the search for a genetic cause of disease. Panel testing of multiple genes has emerged as the preferred approach. However, the method of panel testing presupposes that the abnormalities that are of clinical relevance are confined to the panel of tested genes. Therefore, NGS-based whole-genome sequencing (WGS) and targeted gene panel analysis in combination with complementary methods provide a comprehensive and feasible approach for genetic diagnostics.
We analysed 26 specific genes that are known to underlie aortic aneurysm and dissection in 100 Chinese patients with sporadic STAAD and used WGS to clarify whether genetic variants in suspicious pathogenic genes were associated with sporadic STAAD.
We present the following article in accordance with the STROBE reporting checklist (available at https://dx.doi.org/10.21037/jtd-20-2758).
Methods
Study population and data collection
Patients with suspected STAAD who were admitted to the emergency centre of Fuwai Hospital from 2012 to 2014 were primarily enrolled when blood samples for genetic testing were obtained within 24 hours of admission. Another 568 healthy control samples were obtained from individuals undergoing physical examination. The diagnosis of STAAD was confirmed using multidetector computed tomography scanning. All of the included patients reported the absence of a first-degree relative with aortic aneurysm or dissection in a detailed medical history inquiry. Patients with AD secondary to surgery, trauma and pregnancy were excluded. Baseline characteristic data were recorded, including sex, age, and previous medical histories, such as hypertension, diabetes mellitus, coronary artery disease, smoking status and drinking status. Other recorded clinical characteristics included baseline vital signs at admission (systolic/diastolic blood pressure and heart rate), imaging examinations and hospital management (medical therapy or surgical intervention). An experienced surgeon-in-charge determined the rationale and strategy of the surgical techniques according to the guidelines for the diagnosis and treatment of aortic disease (8,9). The primary end point was in-hospital all-cause mortality, and the evaluation of mortality was obtained from our hospital’s medical database. The study was performed in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the Ethics Committee of Fuwai Hospital (No. 2012-396). Informed consent was obtained from all patients.
WGS and variant calling method
Genomic DNA was isolated from blood samples. Novogene performed WGS. DNA libraries were sequenced on an Illumina HiSeq X according to the manufacturer’s instructions to generate paired-end 150 bp reads, and the researchers were blinded to phenotypic labels during the WGS process. Primer sequences were trimmed from FASTQ files using cutadapt (v 1.9.1) 20 prior to read mapping to the reference genome (UCSC hg19) using BWA-MEM. SAMtools (version 1.0) was used for variant calling and the identification of single-nucleotide variants (SNVs) and indels. Only variants with QualByDepth (QUAL) >20, Depth (DP) >4, and RMS MappingQuality (MQ) >40 passed the filter.
Mutation analysis
WGS was performed to scan for genetic variants that may underlie STAAD. This study also analysed 26 specific previously known genes (with exact OMIM numbers) that underlie aortic aneurysm and dissection. The detailed panel of tested genes is presented in Table 1. Polymorphic variants were excluded if their allelic frequency was >0.01 in the 1000 Genomes Project (in all populations, http://www.internationalgenome.org/1000-genomes-browsers) or in the 568 healthy controls. Variants were considered pathological if they met one of the following criteria: (I) previously reported as pathological in the NCBI ClinVar database; (II) nonsense and indel (frameshift or nonframeshift) mutations; (III) novel missense mutations that indicated a damaging effect in SIFT20 (http://sift.jcvi.org/) or PolyPhen-219 (http://genetics.bwh.harvard.edu/pph2/); and (IV) variations in the splice site within 3 bp of the exon.
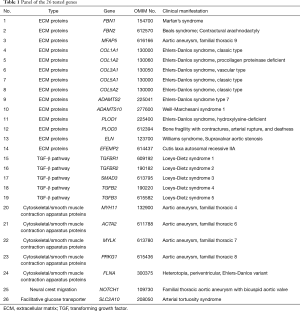
Full table
Statistical analysis
All statistical analyses were performed using SPSS version 19.0 (SPSS, Inc., Chicago, Illinois, USA). Continuous variables are presented as the means ± SD or the medians and interquartile range-based Gaussian distribution. Baseline characteristics were compared between groups using unpaired Student’s t tests or chi-square tests. The in-hospital mortality was compared between the different groups using chi-square tests. A P value of <0.05 was considered statistically significant.
Results
Patient clinical characteristics
In total, 104 sporadic subjects who were diagnosed with STAAD were primarily enrolled, and WGS was performed in 96.1% (100/104) of patients with higher-quality blood DNA samples. The summarized clinical characteristics are shown in Table 2. The average age of these patients was 52.7±12.3 years, and 62.0% (62/100) of the enrolled patients were male. Eighty-four cases showed the DeBakey I AD phenotype, 16 cases were DeBakey type II, and 65 patients (65.0%) had a history of hypertension.
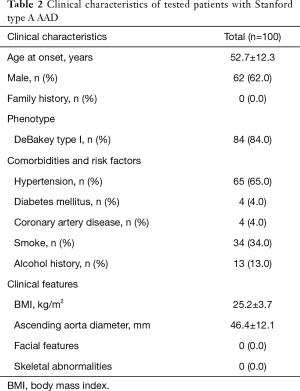
Full table
Technical performance: coverage and variant calling
After the entire run was completed, on average, 89.79 GB of Illumina sequencing data per subject were generated. The average sequencing depth of WGS was 32.58±2.59×, with an average coverage of 99.39% of the genome. The average depth of the analysed target genes of all samples was over 20×, and the coverage of the analysed genes in a specific sample was over 99.38% (Table S1). The percentage of each gene sequence covered using this assay is presented in Table S2. A summary of the quality control for WGS is presented in Table S3.
Genetic variants of panel genes for WGS
Site-based data
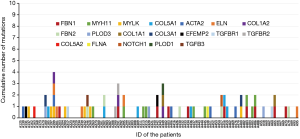
In total, 130 mutations were screened in the tested genes from the panel, including 5 indel mutations and 125 SNV mutations. Seventy mutations were excluded as common polymorphisms or neutral rare variants according to the exclusion criteria. After exclusion, 60 mutations, including 5 indels and 55 SNVs in 19 panel genes, were identified as disease-associated mutations (Figure 1), and the allelic frequency of the identified variants in healthy controls is detailed in Table S4. Fifty-six mutations were novel, and 4 mutations were previously reported. Mutations located in the extracellular matrix (ECM)-coding genes, especially in FBN1 (10 mutations, 16.7%) and COL5A1 (6 mutations, 10.0%), constituted 61.7% (37/60) of these variants. Seventeen mutations (28.3%) were found in cytoskeletal or smooth muscle contraction apparatus protein-coding genes, and 5 mutations were found in coding genes in the transforming growth factor-β (TGF-β) pathway (2 in TGFBR1, 2 in TGFBR2 and 1 in TGFB3). Only 1 mutation was identified in the NOTCH1 gene, which was related to neural crest migration. Detailed site-based information is shown in Table 3.
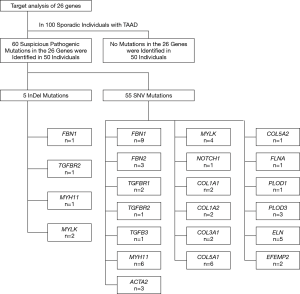
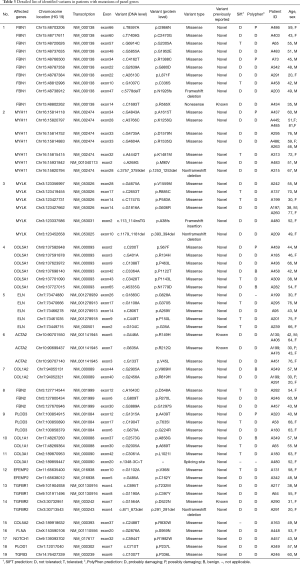
Full table
Case-based data
We identified genetic mutations in half of the patients (50/100) using the gene panel for aortic aneurysm and dissection. The other half of the patients showed no deleterious mutations. From the data, more than 1 variant was found in 14 patients (1 patient had 4 mutations, 2 patients had 3 mutations, and 11 patients had 2 mutations), and the other 36 patients carried only 1 mutation (Figure 2).
Genetic mutations and clinical features and in-hospital death
Baseline clinical feature
Patients were grouped according to the presence of pathogenic variants [with (n=50) or without mutations of the panel genes (n=50)]. Patients with mutations were subdivided into a single-mutation group (n=36) or multiple-mutation group (n=14) according to the number of variants. Patients with mutations were further subdivided into groups based on whether they had mutations in the ECM coding gene (n=29) or only carried mutations in other genes (n=21). Comparisons of clinical characteristics in patients with respect to the presence and type of pathogenic mutations are detailed in Table 4.
Full table
An extended ascending aortic diameter was found in patients with mutations in the panel genes (49.1±12.3 vs. 43.7±11.2 mm, P=0.023) compared to patients without mutations. The DeBakey type I phenotype was more common in patients with mutations in ECM coding genes than in patients with mutations in other genes (96.6% vs. 66.7%, P=0.007). Other clinical features were comparable between the groups (all Ps >0.05). Table S5 shows the clinical characteristics of patients with the top 3 most frequent gene mutations (FBN1, MYH11, MYLK) in this cohort. FBN1 was the most frequently mutated gene (n=10), and the onset age differed significantly between the three genes (P=0.021). The onset age of patients with mutations in FBN1 was younger than that of patients without FBN1 mutations (44.7±11.0 vs. 53.5±12.1, P=0.030).
In-hospital outcome
The overall in-hospital mortality was 12.0% (12/100) in all enrolled patients with STAAD. Of these patients, 66.6% (8/12) died of aortic rupture, and 33.3% (4/12) died of cardiac issues (Table S6). Clinical outcomes in these patients were summarized according to the presence, number and type of mutations (Figure 3). In-hospital mortality was 3-fold higher in patients with mutations than in patients without mutations, but the difference was not statistically significant (18.0% vs. 6.0%, P=0.065). The in-hospital death rate was comparable regardless of the number or type of mutation the patients carried (all Ps >0.05, Figure 3A).
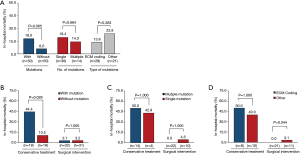
Because of the significant impact of surgical treatment on in-hospital death from STAAD, patients were subdivided into a conservative treatment group and a surgical intervention group. The in-hospital mortality was 27.0% (10/37) and 3.2% (2/63) in patients with conservative and surgical treatments, respectively. Subgroup analysis of in-hospital mortality was also performed in different treatment groups according to the presence, number and type of mutations. Figure 3B shows that in-hospital mortality was comparable between patients with and without mutations who received surgical treatment (3.1% vs. 3.2%, P=1.000). However, increased in-hospital mortality in mutation carriers was observed only in patients who received conservative treatment (44.4% vs. 10.5%, P=0.029). When the in-hospital mortality was compared between the mutation number and mutations in different gene groups, no statistically significant differences were observed (all Ps >0.05, in Figure 3C,D).
Discussion
The current study analysed genetic variants of 26 panel genes that cause thoracic aortic aneurysm and aortic dissection (TAAD) in 100 patients with sporadic STAAD using WGS. Sixty probable disease-causing mutations were identified in half of the enrolled patients for 19 of the 26 panel genes. Fifty-six of these mutations were newly discovered, and 4 mutations were previously reported. Patients with mutations in the panel genes had an ascending aorta with a larger diameter and a poorer in-hospital outcome than patients without mutations. Patients with mutations in ECM coding genes seemed more likely to develop a severe AD phenotype (DeBakey type I) than patients with mutations in other genes.
Previous studies have shown that genetic disorders are associated with aortic aneurysm and dissection. NGS-based gene panel testing is widely used to detect genetic susceptibility to aortic disorders. Three studies investigated the genetic variants in TAAD in European populations. Poninska et al. used whole-exome sequencing (WES) and a gene panel to study 51 patients with TAAD and reported a 35.3% diagnostic yield (10). Campens et al. found variants in 13% of TAAD patients using a panel of 7 genes (11). Proost et al. screened 14 genes from 55 patients and identified 15 pathogenic mutations and six variants of uncertain significance (12). An American study with a larger sample size (n=102) showed that 4.9% of patients carried a pathogenic/likely pathogenic variant, and 22% had a variant of uncertain significance based on a 21-gene panel (13). Wooderchak et al. found pathogenic variants in 10% of patients and variants of uncertain significance in 18% of patients (14). Zheng et al. found genotype-positive variants in 28.8% of TAAD patients in a South Chinese Han cohort using a 69-gene panel (15), and Fang et al. identified 40 variants (3 pathogenic, 10 likely pathogenic and 27 variants of uncertain significance) in 36 of 70 TAAD patients (16). Similar to the two Chinese investigations, the genotype-positive rate in our study was clearly higher than that in Western cohorts. The higher mutation rate may be due to the following reasons. First, we only included patients without a family history, and the genetic changes in sporadic patients may differ from those in patients with a family history. These patients may have a greater likelihood of carrying a single mutation with a relatively lower penetrance and show a sporadic form. Second, we only included patients with STAAD who had a stronger genetic component than patients with Stanford type B AD or patients with aortic aneurysms who showed associations with lifestyle-linked risk factors. Third, the patients included in this study were primarily northern Han Chinese people, whose genetic backgrounds are completely different and who have a younger onset age of AD compared to Western populations.
MFS is the most common aortic aneurysm syndrome caused by heterozygous mutations in FBN1. A genome-wide association study (GWAS) analysed 765 sporadic cases of TAAD, including STAAD, and found that common single-nucleotide polymorphisms (SNPs) in the FBN1 gene were significantly associated with an increased risk of developing aortic aneurysms and dissections (17). Rare mutations of FBN1 were also found in 15.75% of STAAD cases in a Chinese population (18). Mutations in FBN1 were also the most frequent (10 mutations, 1 known and 9 novel) in sporadic STAAD in our study. However, except for a previously reported stop-gain mutation (exon 14, pR565X) (19), none of these mutations were located in the central coding sequences (exons 24–32) of the FBN1 gene, the disturbance of which may cause a severe phenotype of MFS (20,21). The location of these variants may partially explain the milder penetrance and variable expressivity in the sporadic form. Consistent with studies that reported on Marfan and non-Marfan patients with aortic disorders (22,23), patients carrying mutations in FBN1 were younger than other patients in our study.
Collagens are the most important components of the ECM, and certain coding genes in collagens are related to EDS. The syndrome, which causes a disorder in the connective tissue, is divided into different phenotypes based on changes in different genes (24,25). AD is one of the most severe complications of this syndrome, especially in EDS type IV (also known as vascular type, with mutations in COL3A1) (26). Patients with EDS show poor prognosis due to the fragility of aortic tissues and poor wound healing. According to data from a previous study (27) and our study, a considerable proportion of sporadic cases of AD showed likely pathogenic variants in EDS-related genes, which may partially explain the poor outcome in some of the AD patients. Other genetic variants were also found in genes responsible for LDS (mutations related to the TGF-β signalling pathway) at a relatively lower ratio (5% cases). De novo mutations were found in 75% of patients with LDS (28), and LDS can occur in sporadic form (29). Five probable pathogenic variants were also found in LDS-related genes in our cohort, including TGFBR1, TGFBR2 and TGFB3.
The ECM is the key structural component of the aorta, as ECM elements provide elasticity and tensile strength to blood vessel walls. The ECM also provides important growth factors, such as TGF-β (6). In our study, the DeBakey type I phenotype was more common in patients with mutations in ECM coding genes (96.6%), which indicates a larger extent of damage. The findings indicated that defects in the ECM components likely had a destructive impact on the structure of the aorta and showed a distinct relationship with the clinical phenotype.
Genetic variants related to vascular smooth muscle cell (SMC) contractility often show associations with nonsyndromic aortic aneurysms and dissections, such as actin alpha 2 (ACTA2), MYH11 and myosin light-chain kinase (MYLK). The ACTA2 gene encodes an SMC-specific isoform of the contractile protein α-actin (30-32). Mutations in ACTA2 are the most common reason for nonsyndromic aortic aneurysms and dissection and account for approximately 2–4% of sporadic cases of aortic disorder (7,32). Similar to previously reported results, we found that 5% (5/100) of patients carried 3 mutations in ACTA2. Two of these mutations were previously reported (33) but corresponded to different changes (p. R149H and p.R212Q). The number of mutations in MYLK and MYH11 was 13, affecting 16 patients, and all of these mutations were novel. Mutations in MYH11 are associated with familial TAAD (5), but the average age of onset was difficult to judge in different familial forms. Nine sporadic patients who carried mutations in MYH11 in our study were older than the other patients. However, this observation may be due to the limited sample size, and further confirmation in a larger cohort is needed in future studies.
The clinical data of this study revealed that genetic differences showed an association with the phenotype of the individual and clinical outcomes. Variants that increase susceptibility to AD may also increase the risk of in-hospital death. This phenomenon stresses the importance of genetically personalized care and subsequent precision treatment in STAAD, especially patients with mild-to-moderate dilation of the ascending aorta, even without a family history. Although additional novel genes were associated with aortic diseases, routine genetic testing using classic gene panels showed strength in the identification of pathogenic variants in STAAD patients (13). A considerable proportion of STAAD patients were associated with mutations in known pathogenic genes. We also recommend that routine genetic testing be performed first in clinical practice, and other patients with AD may undergo WGS or WES to identify previously unreported changes.
The present study has two important strengths. First, the present study used an enlarged panel of classic pathogenic genes to investigate AD-related genetic changes in a Chinese population with a rare mutation analysis strategy and did not analyse only common polymorphisms. The rate of mutation carriers in our cohort was almost twice the rate of TAAD in a study performed in a Western population (10-14). This difference may partially explain the younger onset age of AD in Chinese patients (34). Second, the present study revealed the potential impact of certain genetic changes on the clinical features and outcomes of AD, which may be of great transitional significance in clinical management.
Several limitations of the present study must be mentioned. First, confounding factors may exist due to the single-centre setting, and the findings obtained in this cohort may not extend to other populations. The small sample size also did not allow for extended analyses of subgroups and corrections. Second, whether the probable pathogenic variants are disease-causing or benign must be determined. Finally, we only primarily reviewed part of the data from WGS, and further analyses and confirmations have not been completed. Despite the relatively small sample size, the clinical features and outcomes showed a relationship with the pathogenic genotype. However, the insufficient sample size restricted further analyses of clinical data, and confirmations from a large multicentre cohort may be needed in the future.
Conclusions
Our study indicated that half of Chinese patients with sporadic STAAD may carry mutations in known pathogenic genes of aortic disease and may exhibit severe clinical features and poor clinical outcomes with conservative treatment.
Acknowledgments
We sincerely appreciate the contributions of the patients and clinic staff who participated in the study. Moreover, the authors acknowledge the effort made by American Journal Experts (
Funding: This work was supported by a grant from the National Natural Science Foundation of China (81570430 to XH Fan, 81470541 to J Cai.), the Open Foundation from Beijing Key Laboratory of Hypertension Research (to XH Fan), and the CAMS Innovation Fund for Medical Sciences (CIFMS, 2016-12 M-1-006 to J Cai).
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at https://dx.doi.org/10.21037/jtd-20-2758
Data Sharing Statement: Available at https://dx.doi.org/10.21037/jtd-20-2758
Peer Review File: Available at https://dx.doi.org/10.21037/jtd-20-2758
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/jtd-20-2758). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was performed in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the Ethics Committee of Fuwai Hospital (No. 2012-396). Informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hagan PG, Nienaber CA, Isselbacher EM, et al. The International Registry of Acute Aortic Dissection (IRAD): new insights into an old disease. JAMA 2000;283:897-903. [Crossref] [PubMed]
- Mussa FF, Horton JD, Moridzadeh R, et al. Acute Aortic Dissection and Intramural Hematoma: A Systematic Review. JAMA 2016;316:754-63. [Crossref] [PubMed]
- Isselbacher EM, Lino Cardenas CL, Lindsay ME. Hereditary Influence in Thoracic Aortic Aneurysm and Dissection. Circulation 2016;133:2516-28. [Crossref] [PubMed]
- van ‘t Hof FN, Ruigrok YM, Baas AF, et al. Impact of inherited genetic variants associated with lipid profile, hypertension, and coronary artery disease on the risk of intracranial and abdominal aortic aneurysms. Circ Cardiovasc Genet 2013;6:264-70. [Crossref] [PubMed]
- Albornoz G, Coady MA, Roberts M, et al. Familial thoracic aortic aneurysms and dissections--incidence, modes of inheritance, and phenotypic patterns. Ann Thorac Surg 2006;82:1400-5. [Crossref] [PubMed]
- Goldfinger JZ, Halperin JL, Marin ML, et al. Thoracic aortic aneurysm and dissection. J Am Coll Cardiol 2014;64:1725-39. [Crossref] [PubMed]
- Pomianowski P, Elefteriades JA. The genetics and genomics of thoracic aortic disease. Ann Cardiothorac Surg 2013;2:271-9. [PubMed]
- Hiratzka LF, Bakris GL, Beckman JA, et al. 2010 ACCF/AHA/AATS/ACR/ASA/SCA/SCAI/SIR/STS/SVM Guidelines for the diagnosis and management of patients with thoracic aortic disease. A Report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines, American Association for Thoracic Surgery, American College of Radiology,American Stroke Association, Society of Cardiovascular Anesthesiologists, Society for Cardiovascular Angiography and Interventions, Society of Interventional Radiology, Society of Thoracic Surgeons,and Society for Vascular Medicine. J Am Coll Cardiol 2010;55:e27-e129. [Crossref] [PubMed]
- Erbel R, Aboyans V, Boileau C, et al. 2014 ESC Guidelines on the diagnosis and treatment of aortic diseases: Document covering acute and chronic aortic diseases of the thoracic and abdominal aorta of the adult. The Task Force for the Diagnosis and Treatment of Aortic Diseases of the European Society of Cardiology (ESC). Eur Heart J 2014;35:2873-926. [Crossref] [PubMed]
- Poninska JK, Bilinska ZT, Franaszczyk M, et al. Next-generation sequencing for diagnosis of thoracic aortic aneurysms and dissections: diagnostic yield, novel mutations and genotype phenotype correlations. J Transl Med 2016;14:115. [Crossref] [PubMed]
- Campens L, Callewaert B, Muino Mosquera L, et al. Gene panel sequencing in heritable thoracic aortic disorders and related entities - results of comprehensive testing in a cohort of 264 patients. Orphanet J Rare Dis 2015;10:9. [Crossref] [PubMed]
- Proost D, Vandeweyer G, Meester JA, et al. Performant Mutation Identification Using Targeted Next-Generation Sequencing of 14 Thoracic Aortic Aneurysm Genes. Hum Mutat 2015;36:808-14. [Crossref] [PubMed]
- Ziganshin BA, Bailey AE, Coons C, et al. Routine Genetic Testing for Thoracic Aortic Aneurysm and Dissection in a Clinical Setting. Ann Thorac Surg 2015;100:1604-11. [Crossref] [PubMed]
- Weerakkody R, Ross D, Parry DA, et al. Targeted genetic analysis in a large cohort of familial and sporadic cases of aneurysm or dissection of the thoracic aorta. Genet Med 2018;20:1414-22. [Crossref] [PubMed]
- Zheng J, Guo J, Huang L, et al. Genetic diagnosis of acute aortic dissection in South China Han population using next-generation sequencing. Int J Legal Med 2018;132:1273-80. [Crossref] [PubMed]
- Fang M, Yu C, Chen S, et al. Identification of Novel Clinically Relevant Variants in 70 Southern Chinese patients with Thoracic Aortic Aneurysm and Dissection by Next-generation Sequencing. Sci Rep 2017;7:10035. [Crossref] [PubMed]
- LeMaire SA, McDonald ML, Guo DC, et al. Genome-wide association study identifies a susceptibility locus for thoracic aortic aneurysms and aortic dissections spanning FBN1 at 15q21.1. Nat Genet 2011;43:996-1000. [Crossref] [PubMed]
- Guo J, Cai L, Jia L, et al. Wide mutation spectrum and frequent variant Ala27Thr of FBN1 identified in a large cohort of Chinese patients with sporadic TAAD. Sci Rep 2015;5:13115. [Crossref] [PubMed]
- Youil R, Toner TJ, Bull E, et al. Enzymatic mutation detection (EMD) of novel mutations (R565X and R1523X) in the FBN1 gene of patients with Marfan syndrome using T4 endonuclease VII. Hum Mutat 2000;16:92-3. [Crossref] [PubMed]
- Halliday DJ, Hutchinson S, Lonie L, et al. Twelve novel FBN1 mutations in Marfan syndrome and Marfan related phenotypes test the feasibility of FBN1 mutation testing in clinical practice. J Med Genet 2002;39:589-93. [Crossref] [PubMed]
- Bunton TE, Biery NJ, Myers L, et al. Phenotypic alteration of vascular smooth muscle cells precedes elastolysis in a mouse model of Marfan syndrome. Circ Res 2001;88:37-43. [Crossref] [PubMed]
- Tsai SH, Lin YY, Hsu CW, et al. The characteristics of acute aortic dissection among young Chinese patients: a comparison between Marfan syndrome and non-Marfan syndrome patients. Yonsei Med J 2009;50:239-44. [Crossref] [PubMed]
- Ueda T, Shimizu H, Aeba R, et al. Prognosis of Marfan and non-Marfan patients with cystic medial necrosis of the aorta. Jpn J Thorac Cardiovasc Surg 1999;47:73-8. [Crossref] [PubMed]
- Byers PH, Murray ML. Heritable collagen disorders: the paradigm of the Ehlers-Danlos syndrome. J Invest Dermatol 2012;132:E6-11. [Crossref]
- Malfait F, Wenstrup RJ, De Paepe A. Clinical and genetic aspects of Ehlers-Danlos syndrome, classic type. Genet Med 2010;12:597-605. [Crossref] [PubMed]
- Byers PH. Ehlers-Danlos syndrome type IV: a genetic disorder in many guises. J Invest Dermatol 1995;105:311-3. [Crossref] [PubMed]
- Li Z, Zhou C, Tan L, et al. Variants of genes encoding collagens and matrix metalloproteinase system increased the risk of aortic dissection. Sci China Life Sci 2017;60:57-65. [Crossref] [PubMed]
- Jones JA, Spinale FG, Ikonomidis JS. Transforming growth factor-beta signaling in thoracic aortic aneurysm development: a paradox in pathogenesis. J Vasc Res 2009;46:119-37. [Crossref] [PubMed]
- Ha JS, Kim YH. A sporadic case of Loeys-Dietz syndrome type I with two novel mutations of the TGFBR2 gene. Korean J Pediatr 2011;54:272-5. [Crossref] [PubMed]
- Disabella E, Grasso M, Gambarin FI, et al. Risk of dissection in thoracic aneurysms associated with mutations of smooth muscle alpha-actin 2 (ACTA2). Heart 2011;97:321-6. [Crossref] [PubMed]
- Zhu L, Bonnet D, Boussion M, et al. Investigation of the MYH11 gene in sporadic patients with an isolated persistently patent arterial duct. Cardiol Young 2007;17:666-72. [Crossref] [PubMed]
- Guo DC, Pannu H, Tran-Fadulu V, et al. Mutations in smooth muscle alpha-actin (ACTA2) lead to thoracic aortic aneurysms and dissections. Nat Genet 2007;39:1488-93. [Crossref] [PubMed]
- Morisaki H, Akutsu K, Ogino H, et al. Mutation of ACTA2 gene as an important cause of familial and nonfamilial nonsyndromatic thoracic aortic aneurysm and/or dissection (TAAD). Hum Mutat 2009;30:1406-11. [Crossref] [PubMed]
- Wang W, Duan W, Xue Y, et al. Clinical features of acute aortic dissection from the Registry of Aortic Dissection in China. J Thorac Cardiovasc Surg 2014;148:2995-3000. [Crossref] [PubMed]


