
Altered gut microbiome compositions are associated with the severity of asthma
Introduction
Asthma is a common respiratory disease, which is characterized by chronic inflammation. This condition affects approximately 1–18% of the population in different countries via both complicated etiopathogeneses and unknown pathogeneses (1). Recently, there has been a research focus on microbiota for its potential roles in patients with asthma. This study, as well as other studies, has shown the disordered lung microbiota and its contributions to the severity and phenotypes of asthma (2-7).
In addition to lung microbiota, gut microbiome diversity has been implicated in the pathogenesis of asthma (8-11). The population diversity of intestinal bacteria is closely related to the microenvironment and immune system. Gut microbiota activate immune responses at distal sites (e.g., the lungs) by secreting factors, such as histamine and short-chain fatty acids (SCFAs) (12,13). Early life and eating habits can also affect the population diversity of intestinal bacteria. For example, excess drinks with free fructose probably lead to higher fructose/glucose ratios in the gut, which weakens intestinal absorption of fructose. The variation of microenvironment shifts the diversity of intestinal bacteria, which finally increases the incidence of asthma (14,15). Alterations in the gut microbiota early in life are associated with the risk of asthma (16-23). However, the precise underlying mechanisms of how gut microbiota affect the pathogenesis of asthma remain unknown, especially for severe asthma.
Moreover, it is unknown whether the severity and/or clinical phenotypes of asthma are linked to different profiles of gut microbiota. Therefore, in this study, we assessed 44 fecal samples from patients with and without asthma in the subtropical city of Guangzhou, within Southern China. High-throughput 454 sequencing was utilized to evaluate fecal samples from 15 severe asthma (SA), 14 non-severe asthma (NSA), and 15 healthy subjects.
We present the following article in accordance with the STROBE Checklist (available at https://dx.doi.org/10.21037/jtd-20-2189).
Methods
Subjects
In this study, 29 outpatients with physician-diagnosed asthma and 15 healthy subjects were recruited at The First Affiliated Hospital of Guangzhou Medical University between December 2016 and June 2017. In patients with asthma 15 were classified as having SA and 14 were classified as having NSA. The proportions of smokers were similar in both the SA (1/15) and NSA (1/14) groups, and there were no smokers in the healthy group. The diagnoses and severities of asthma were confirmed based on the criteria of the Global Initiative for Asthma (GINA) (1), while severe asthma was defined in accordance with the European Respiratory Society/American Thoracic Society (ERS/ATS) guidelines (24). We grouped these patients with asthma into the following two phenotypes on the basis of total immunoglobulin E (IgE) levels: the high-IgE asthma (HEA; >60 IU/mL) group and the low IgE asthma (LEA; <60 IU/mL) group. The exclusion criteria considered for this study included the following: subjects diagnosed with other lung diseases (chronic obstructive pulmonary disease, bronchiectasis and interstitial pneumonia), auto-immune diseases (inflammatory bowel disease), or other diseases known to affect the diagnosis of asthma. Patients taking antibiotics at the time of this study or those receiving immunosuppressive therapies and/or allergen-specific immunotherapies within the last year were also excluded from the study. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Written informed consent was obtained from all subjects and the research was approved by the ethics committee of the First Affiliated Hospital of Guangzhou Medical University (approval No.2016-68).
Atopy
Levels of specific IgE to common aeroallergens (Hx2 and Mx2) and the food allergen (Fx5) were determined using the ImmunoCAP system (Thermo Scientific, Uppsala, Sweden). Specific IgE was defined as positive if the measurement was ≥0.35 IU/mL. Subjects were characterized as atopic if they had at least one positive specific IgE.
Collection and processing of fecal samples
Fresh fecal samples (1–2 g per person) were collected and transferred from bedpans that had been disinfected with 75% ethanol. The fecal samples were then manipulated and homogenized. The prepared samples were then collected and stored at −80 °C until further use (11).
DNA extraction and sequencing
The fecal samples (10–50 g) were collected with Tris-EDTA buffer (10 mMTris-HCl, 1 M EDTA, pH 8.0). Bacterial DNA was isolated and purified using a NEBnext microbiome DNA enrichment kit (New England Biolabs, Ipswich, MA, USA) in line with the manufacturer’s instructions. A standard concentration of 10 ng/mL of DNA was prepared for each individual sample for all PCR assays. After performing a sample quality test, we used those quantified DNA samples to construct a library. The bacterial DNA was amplified for the V4 hypervariable region of bacterial 16s rRNA. PCR was amplified with the fusion primer; then, short fragments were removed using Ampure beads. Finally, the quantified library was used for sequencing with the Illumina Miseq 2500 platform (BGI, Shenzhen, China).
Statistical analysis
Reads that did not match the barcode or the primer sequences were excluded before data analysis. Unique reads were pre-clustered by scripts written in Mothur (version 1.31.2, http://www.mothur.org/), within two mismatch similarities. Operational taxonomic unit (OTU) numbers were assessed by scripts written in Mothur based on 97% similarities. First, unique reads were aligned to the Ribosome Database Project database using scripts of software written in Mothur and were subsequently annotated. Furthermore, when more than 51% of unique reads of the OTUs were interpreted to belong to one species, we defined these OTUs as representing a unique species. Then, OTUs were calculated by taxonomic analysis at a higher level, such as from the genus level, to the family level, and were summarized using Mothur software. To determine abundance information of the fecal samples, the numbers of reads in each OTU were computed. The distribution of all annotated species or OTUs at different taxonomic levels (i.e., phylum, class, order, family, and genus) among different samples was analyzed and summarized by scripts of software written in QIIME (version 1.50). We not only used the Chao1 estimator and ACE to estimate species richness but also used Shannon-Wiener and Simpson indices to assess the ecological diversity of various species. Differences in bacterial abundances and clinical parameters between healthy and asthmatic subjects were assessed using analysis of variance (ANOVA). We performed linear correlations to determine associations among clinical and demographical features and bacterial abundances in healthy subjects and patients with asthma. We selected 30 variables in cluster analysis; 22 variables were excluded as being clinically irrelevant for the clustering and being represented by other variables. Eventually, eight variables were selected. The SPSS statistical software package (version 18.0) was used for all statistical analyses. A P value <0.05 was considered to be statistically significant. Data were reported as the mean ± standard deviation or median with interquartile range. Analysis of LSD, Bonferroni, and Spearman’s rank correlation coefficient was used for parametric continuous data; analysis of Kruskal–Wallis, Mann-Whitney U, and Pearson’s correlation was used for nonparametric continuous data; and the chi-square test was used for categorical variables.
Results
Sociodemographic characteristics of the included subjects
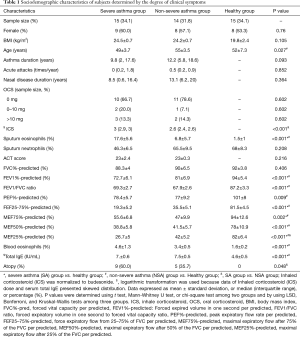
Sociodemographic and clinical features of the subjects are presented in Table 1. There were no significant differences in gender or body mass index (BMI) among the three groups: patients with SA, referred as the SA group, patients with NSA, referred as the NSA group, and the healthy subjects, referred as the healthy group, in this article. Compared with the healthy group, the SA and NSA groups showed higher eosinophils in both sputum and blood, suggestive of eosinophilic-predominant airway inflammation. Compared with the NSA group, the SA group had a higher dose of inhaled corticosteroids (ICS) and a higher percentage of atopy (60% vs. 35.7%, respectively; P<0.05).
Full table
Compositions of gut microbiota
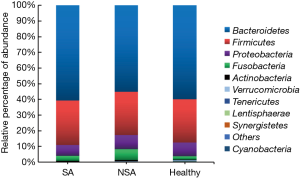
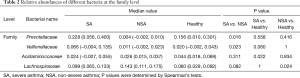
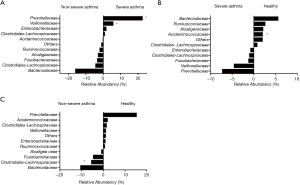
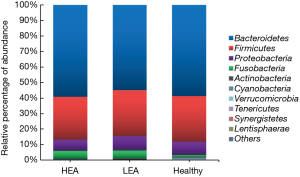
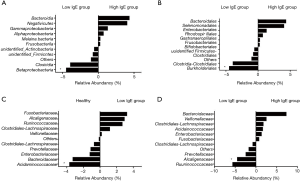
We assessed bacterial richness by OTU numbers and evaluated species diversity by Chao, ACE, Shannon, and Simpson indices. In this study, a total of 3,274,052 raw reads were generated for all samples, of which 2,651,837 quality sequence reads were used for subsequent OTU analyses. We found no statistically significant differences among the three groups in the number of OTU, Chao, ACE, Shannon, or Simpson indices, indicating that gut microbial richness and diversity were quite equal among the three groups. We also identified 11 bacterial phyla in all of the fecal samples, of which Bacteroidetes, Proteobacteria, Firmicutes, and Fusobacteria contributed to over 90% of the total sequences in all three groups (Figure 1). There was no significant difference in the relative abundance of bacteria at the phylum level among the three groups. However, at the family level, we confirmed that the relative abundance of Acidaminococcaceae was remarkably lower in the SA group, than that in the healthy subjects (P<0.05). Lachnospiraceae had a higher proportion in the NSA group than in the healthy group (P<0.05). Veillonellaceae and Prevotellaceae were observably more common in samples from the SA group than those from the NSA group (P<0.05) (Table 2 and Figure 2).
Full table
The association of gut microbiota and severity of asthma
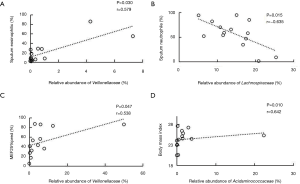
Analysis based on the family level showed that the relative abundance of Veillonellaceae was positively related to the proportion of sputum eosinophilia in the NSA group (r=0.579, P=0.030) (Figure 3A). In contrast, the relative abundance of Lachnospiraceae was negatively correlated with sputum neutrophils (%) in the NSA group (r=−0.635, P=0.015) (Figure 3B). Additionally, positive associations were observed not only between the relative abundance of Veillonellaceae and MEF25% (Figure 3C), but also between that of Acidaminococcaceae and BMI (r=0.642, P=0.010) (Figure 3D) in the SA group.
Differential gut microbiome profiles according to the level of total IgE
In view of the potential role of total IgE levels in the gut microbiome profiles, we grouped our patients into the high-IgE asthma (HEA) group, low-IgE asthma (LEA) group, and healthy group. As shown in Table 3, there were 21 subjects (72.4% of all of the asthmatic patients) in the HEA group. Among them, 11 cases belonged to the SA group and 10 cases to the NSA group. Compared with the LEA group, acute attacks were obviously elevated in the HEA group. No statistical differences in OTU numbers, and Chao, ACE, Shannon, and Simpson indices were observed among the three groups. Similar to the SA and NSA groups, a core gut microbiome consisting of Bacteroidetes, Proteobacteria, Firmicutes, and Fusobacteria at the phylum level was identified in the HEA and LEA groups (Figure 4). No significant differences in the relative abundance of bacteria at the phylum level were detected among the three groups. However, at the class level, Betaproteobacteria was more frequent in the LEA group than in the HEA group (P=0.018). Moreover, Burkholderiales was significantly more abundant in the LEA group than in the HEA group at the order level (P=0.018). Finally, Alcaligenaceae was confirmed to be more common in the LEA group than in the HEA group at the family level (P=0.019) (Table 4, Figure 5).
Full table
Full table
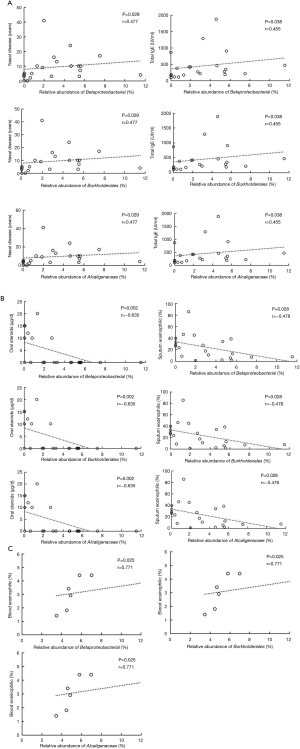
According to correlation analysis, nasal disease duration and serum total IgE showed a positive correlation between the three bacteria levels (Betaproteobacteria class, Burkholderiales order, and Alcaligenaceae family) in the HEA group (P<0.05) (Figure 6A I-VI). In contrast, significant negative correlations were found between the relative abundance of the abovementioned bacteria and the dose of oral steroids and sputum eosinophilia in the HEA group (Figure 6B I-VI). The relative abundance of Betaproteobacteria class, Burkholderiales order, and Alcaligenaceae family showed a positive relationship with the blood eosinophilic (%) (P<0.05) in the LEA group (Figure 6C I-III).
Principal component analysis
Five principal components (PC1-5) with eight values >0.8 and accounting for >10% of the variance were identified (Table 5). PC1 was characterized by severe asthma, which correlated with a worse reaction to ICS and an elevation of sputum neutrophils. Moreover, PC1 was associated with an increased abundance of Acidaminococcaceae. The features of PC2 included longer duration of asthma and nasosinusitis, without associated bacterial species. PC3, which had a lower abundance of Prevotellaceae, was also related to severe asthma. PC4 was grouped into non-severe asthma, with a better reaction to ICS and an increased abundance of Veillonellaceae. PC5 was typified by upregulated sputum neutrophils, reduction of Preveotllaceae abundance, and elevation of Acidaminococcaceae abundance.
Full table
Discussion
Previous studies have shown that intestinal bacteria are closely related to the occurrence and development of asthma in infants and adults (8,11,18,25-29). To our knowledge, a few studies have reported on the relationships of gut microbiota with different levels of severity and serum total IgE in asthma.
This study collected 44 fecal samples (29 subjects with asthma and 15 healthy) to conduct DNA sequencing. Twenty-nine subjects with asthma were grouped into two subgroup based on the inhaled corticosteroid (ICS) dose and Total IgE (Tables 1 and 4). The aim of this study was to explore the relationship between the characteristics of intestinal flora from patients with asthma and asthmatic phenotype.
The diagnosis of severe and non-severe asthma was established based on the criteria of the GINA (1), and European Respiratory Society/American Thoracic Society (ERS/ATS) guidelines (24). Our results showed that four predominate bacterial phyla exist in the gut among the SA, NSA and healthy groups. They included Bacteroidetes, Proteobacteria, Firmicutes, and Fusobacteria (Figure 1). This result was inconsistent with the findings of Hevia et al. (11). and Hua et al. (30). We did not obtain significant alpha-diversity indexes, including Shannon, Simpson, Chao1, and ACE indices, among the SA, NSA, and healthy individuals.
A recent study has suggested that higher relative abundances of airborne Lachnospiraceae and Prevotellaceae lower the prevalence of asthma and allergy (26). Moreover, lower Lachnospiraceae in a three-month infant was identified as a risk factor of an early allergic wheezy phenotype, whereas Veillonellaceae was a protective factor against allergic disease (27). However, in our present study, Lachnospiraceae, Veillonellaceae and Prevotellaceae families were more common in the guts of individuals in the SA and NSA groups (Table 2, Figure 2). We adopted different ways of classification according to the levels of severity of asthma and Total IgE, which caused discrepancy in previous studies. Furthermore, the mode of delivery, antibiotics given to the mother or infant, the mode of infant feeding, environment (31,32), food habits, gene differences (33), and anatomical and physiochemical differences among the intestines (34) influence the establishment of the gut microbiota. The subjects of this study were Chinese, whose environment, food habits, and genes were different from those in European and American subjects. Therefore, the results of this study are helpful to explore the relationship between the diversity of intestinal flora and the development of asthma in different regions of the world.
A previous study suggested that two strains of Veillonella in stool samples of atopic wheezing children (22). Additionally, Arrieta et al. have found that intestinal Prevotellaceae family induces TH2 immune responses (35), which was probably closely related to asthma. Furthermore, some studies have indicated that Lachnospiraceae family play an important role in remolding of airways (36), which was also likely associated with obstructive ventilation dysfunction in asthma patients. The major modulation of intestinal flora on the disease may involve the following mechanisms: First, the molecular structure of MAMPs (including LPS and polysaccharide A) for intestinal flora is recognized by pattern-recognition receptors in humans and activate the nuclear factor kB pathway, thus inducing transcription of proinflammatory and other cytokine genes (37,38). Second, SCFAs, including acetate, propionate, and butyrate, the major end-products metabolized by mainly anaerobic bacteria in the intestine, affect the colonization of other bacteria in the intestine and contribute to maintaining the intestinal epithelium, allergen uptake, and immunological responses in allergic asthma (39). Prevotellaceae family has the structure of MAMPs (LPS) (40,41), and it has the ability to produce SCFAs, as do the Veillonellaceae family, Acidaminococcaceae family, and Lachnospiraceae family (40,41). We speculated that the cellular structure and metabolites of the intestinal flora may be related to asthma severity levels.
To further explore the relationship between different intestinal bacteria and asthma, we took the data of OTUs from intestinal flora and clinical indicators to make the correlation analysis. The relative abundance of Veillonellaceae showed a positive correlation with MEF25% predicted and sputum eosinophils (%) in the SA and NSA groups (Figure 3A,C). Veillonella takes part in atopic wheezing and airway obstructions in children (35) and adults (28). It populates the small intestine and appears to block IL-12p70 production while augmenting IL-8, IL-6, IL-10, and TNF-α responses by dendritic cells in in vitro assays; indeed, these cytokines have been linked to airway obstruction and severe asthma phenotype (41-45), as well as promotion of eosinophil recruitment and airway inflammation (46-48). A growing number of studies have found that the increase of the relative abundance of Lachnospiraceae family inhibits the production of IL-17, the cytokine that may lead to improvement in clinical symptoms of asthma (36,49), and indirectly results in the reduction of neutrophilic ratio in induced sputum (50). This was similar to the results of our study (Figure 3B). Finally, as an opportunistic pathogen of the intestine, genus Acidaminococcus belongs to the family Veillonellaceae, which is known for producing succinate, a potential microbe-derived metabolite associated with an increased risk of cardiovascular disease (51). The function of succinate may be in activating hypoxia-inducible factor HIF-1α, a key transcription factor underlying chronic inflammation and dysfunction in obesity (52). Elevation of BMI has been described as an independent risk factor for adult and pediatric severe asthma (53,54). Additionally, a higher BMI increases the risk of asthma morbidity associated with respiratory tract infections (55). In our study, BMI positively correlated with the relative abundance of Acidaminococcaceae in the SA group (Figure 3D). We speculated that the Acidaminococcaceae family (Firmutes phylum) probably increased the incidence of asthma. The characteristics of severe asthma in our study included the following: impairment of lung function (decreased MEF25% predicted), elevated eosinophil counts (in sputum), and elevated BMI. We considered that the intestinal flora may be closely related to the different levels of severity of asthma, given the correlation between clinical indicators and the relative abundance of intestinal flora.
In terms of the epidemiology and natural history of asthma, the outcomes and treatment regimens (TENOR) study and other studies have elucidated that total serum IgE levels are associated with asthma severity (56,57). At present, the anti-IgE antibody, omalizumab, has been recommended for use as an add-on therapy in severe asthma (9,58). Furthermore, the potential for indoor microbial dysbiosis to contribute to asthma severity is determined by serum IgE levels (59). In support of the hypothesis that the role of serum IgE levels in gut microbiome dysbiosis of asthmatic subjects remains unknown. Our study divided all of the asthmatic patients into the HEA and LEA groups (Table 3). Some studies found that the range of serum total IgE in asthma fluctuated greatly (60-62). It was hard to obtain the accurate levels of serum IgE to define the asthma phenotypes. We selected 60 IU/mL as a cutoff value of serum total IgE on the basis of The Clinical Allergic Diseases (63). The features of the HEA group included poor management of asthma (high risk of exacerbation) and higher eosinophil levels (in sputum and blood). Compared with those in the LEA group, the relative abundances of Betaproteobacteria class, Burkholderiales order and Alcaligenaceae family were significantly low in the HEA group (Figure 5). We also found significant correlations between the bacterial abundances and clinical features in both the HEA and LEA groups (Figure 6).
Serum total immunoglobulin E is secreted by sensitized B-cells; the B-cells binds to an allergen and then to a receptor on mast cells or basophils, where they triggers the release of inflammatory mediators (64). It can be affected by metabolites of intestinal microflora which can maintain the immune regulatory T-cell survival and its function (64). On the one hand, it has been demonstrated that a modulation of HDM exposure (which elevated immunoglobulin E level) in case of microbial dysbiosis can change the gut microbial community (65). In addition, IgE is also produced locally in the gut; the particular subsets of airway microbiome dysbiosis-associated HDM-specific IgE response can interact with corresponding gut microbiome by changing the IgE levels in the gut through the gut-lung axis (65,66). On the other hand, the gut microbiome affects the levels of serum IgE depending on metabolites. Two studies indicated that low intestinal Proteobacteria phylum during the first week and years of life was associated with IgE-associated eczema (67,68). Furthermore, there was a correlation between the decrease in the relative abundance of intestinal Proteobacteria phylum and serum IgE in patients with allergic diseases; the potential mechanism was that the molecular structure and metabolites of the intestinal flora were recognized by receptors TLR driving differentiation of Treg cells, which caused imbalance between Th1 and Th2 immune responses (47,48,67,68) and changed the serum IgE and eosinophils in sputum and blood (47). In contrast, Liu et al. suggested that higher levels of Proteobacteria phylum may lead to an increase in LPS content and increase the risk of inflammation and immunological abnormality, such as allergic rhinitis (69). In general, a potential relationship exists between the relative abundance of Proteobacteria phylum (including Betaproteobacteria class, Burkholderiales order, and Alcaligenaceae family) and serum total IgE from allergy disease; however, further studies are needed to fully understand the relations between gut microbiome dysbiosis and serum IgE levels in patients with asthma.
Finally, the OTUs of intestinal flora were extracted to conduct a principal component analysis with clinical indicators from subjects with asthma. The results showed that a higher dose of ICS was associated with more severe asthma. The implications of Acidaminococcaceae family, Prevotellaceae family, and Veillonellaceae family in specific phenotypes of severe asthma were also revealed by principal component analysis in our study (Table 5). This may help to take the intestinal flora as a biomarker to determine asthmatic phenotype.
This study had limitations. First, the current work was based on a small sample size. Therefore, the effects of gut microbiome dysbiosis in severe asthma need to be further explored in studies with larger sample sizes and with extend duration of observations. Second, several confounding factors have also influenced gut microbiome dysbiosis, such as geographical region, diet, and indoor/outdoor pollution. Third, changes in food composition were associated with intestinal flora disorder, especially the ratio of fructose to glucose (14,15); thus, it would have been valuable to evaluate participants for fructose malabsorption via a hydrogen breath test. Finally, our study did not include paired sputum samples.
Overall, our present study revealed that altered gut microbiome may be associated with the severity of asthma. Furthermore, our findings may aid in further understanding the relationship between serum IgE levels and gut microbiome dysbiosis in asthmatic patients.
Conclusions
Gut microbiome plays an important role in the development of asthma. Altered gut microbiome composition is related to characteristics of asthma, including the severity of asthma and phenotypes defined by serum IgE levels.
Acknowledgments
We would like to thank Dr. Luo Wei and Dr. Chen Qiaoli for the induced sputum tests, Dr. Yu Xinxin, Dr. An Jiaying, and Dr. Liu Wenting for performing the pulmonary function tests, and Dr. Wei Nini for the serum allergy tests.
We appreciate the contribution of investigators who participated in treating asthmatic patients with asthma and their efforts in sample collection and data collection. We thank LetPub (
Funding: The Natural Science Foundation of Guangdong Province (Grant no. 2019A1515010622), The Foundation for High-Level University Construction of Guangzhou Medical University (Professor Kian Fan Chung project, Grant no. B195002010041), and The Precision Medicine Research of the National Key Research and Development Plan of China (Grant no. 2016YFC0905800).
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at https://dx.doi.org/10.21037/jtd-20-2189
Data Sharing Statement: Available at https://dx.doi.org/10.21037/jtd-20-2189
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form. (Available at https://dx.doi.org/10.21037/jtd-20-2189). Prof. Kian Fan Chung serves as an unpaid associate editor-in-chief of Journal of Thoracic Disease. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Written informed consent was obtained from all subjects, and the research was approved by the ethics committee of the First Affiliated Hospital of Guangzhou Medical University (approval No. 2016-68).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bateman ED, Hurd SS, Barnes PJ, et al. Global strategy for asthma management and prevention: GINA executive summary. Eur Respir J 2008;31:143-78. [Crossref] [PubMed]
- Li N, Qiu R, Yang Z, et al. Sputum microbiota in severe asthma patients: Relationship to eosinophilic inflammation. Respir Med 2017;131:192-8. [Crossref] [PubMed]
- Denner DR, Sangwan N, Becker JB, et al. Corticosteroid therapy and airflow obstruction influence the bronchial microbiome, which is distinct from that of bronchoalveolar lavage in asthmatic airways. J Allergy Clin Immunol 2016;137:1398-1405.e3. [Crossref] [PubMed]
- Simpson JL, Daly J, Baines KJ, et al. Airway dysbiosis: Haemophilus influenzae and Tropheryma in poorly controlled asthma. Eur Respir J 2016;47:792-800. [Crossref] [PubMed]
- Zhang Q, Cox M, Liang Z, et al. Airway Microbiota in Severe Asthma and Relationship to Asthma Severity and Phenotypes. PLoS One 2016;11:e0152724 [Crossref] [PubMed]
- Huang YJ, Nariya S, Harris JM, et al. The airway microbiome in patients with severe asthma: Associations with disease features and severity. J Allergy Clin Immunol 2015;136:874-84. [Crossref] [PubMed]
- Green BJ, Wiriyachaiporn S, Grainge C, et al. Potentially pathogenic airway bacteria and neutrophilic inflammation in treatment resistant severe asthma. PLoS One 2014;9:e100645 [Crossref] [PubMed]
- Barcik W, Boutin RCT, Sokolowska M, et al. The Role of Lung and Gut Microbiota in the Pathology of Asthma. Immunity 2020;52:241-55. [Crossref] [PubMed]
- Mavissakalian M, Brady S. The Current State of Biologic Therapies for Treatment of Refractory Asthma. Clin Rev Allergy Immunol 2020;59:195-207. [Crossref] [PubMed]
- Kang YB, Cai Y, Zhang H. Gut microbiota and allergy/asthma: From pathogenesis to new therapeutic strategies. Allergol Immunopathol (Madr) 2017;45:305-9. [Crossref] [PubMed]
- Hevia A, Milani C, López P, et al. Allergic Patients with Long-Term Asthma Display Low Levels of Bifidobacterium adolescentis. PLoS One 2016;11:e0147809 [Crossref] [PubMed]
- Smolinska S, Groeger D, O'Mahony L. Biology of the Microbiome 1: Interactions with the Host Immune Response. Gastroenterol Clin North Am 2017;46:19-35. [Crossref] [PubMed]
- Barcik W, Pugin B, Westermann P, et al. Histamine-secreting microbes are increased in the gut of adult asthma patients. J Allergy Clin Immunol 2016;138:1491-1494.e7. [Crossref] [PubMed]
- DeChristopher LR, Tucker KL. Excess free fructose, apple juice, high fructose corn syrup and childhood asthma risk - the National Children's Study. Nutr J 2020;19:60. [Crossref] [PubMed]
- Ferraris RP, Choe JY, Patel CR. Intestinal Absorption of Fructose. Annu Rev Nutr 2018;38:41-67. [Crossref] [PubMed]
- Frei R, Ferstl R, Roduit C, et al. Exposure to nonmicrobial N-glycolylneuraminic acid protects farmers' children against airway inflammation and colitis. J Allergy Clin Immunol 2018;141:382-390.e7. [Crossref] [PubMed]
- Fujimura KE, Sitarik AR, Havstad S, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med 2016;22:1187-91. [Crossref] [PubMed]
- Huang YJ, Boushey HA. The microbiome in asthma. J Allergy Clin Immunol 2015;135:25-30. [Crossref] [PubMed]
- Roduit C, Frei R, Depner M, et al. Increased food diversity in the first year of life is inversely associated with allergic diseases. J Allergy Clin Immunol 2014;133:1056-64. [Crossref] [PubMed]
- Frei R, Roduit C, Bieli C, et al. Expression of genes related to anti-inflammatory pathways are modified among farmers' children. PLoS One 2014;9:e91097 [Crossref] [PubMed]
- Abrahamsson TR, Jakobsson HE, Andersson AF, et al. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin Exp Allergy 2014;44:842-50. [Crossref] [PubMed]
- van Nimwegen FA, Penders J, Stobberingh EE, et al. Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. J Allergy Clin Immunol 2011;128:948-55.e1-3.
- Braun-Fahrländer C, Riedler J, Herz U, et al. Environmental exposure to endotoxin and its relation to asthma in school-age children. N Engl J Med 2002;347:869-77. [Crossref] [PubMed]
- Chung KF, Wenzel SE, Brozek JL, et al. International ERS/ATS guidelines on definition, evaluation and treatment of severe asthma. Eur Respir J 2014;43:343-73. [Crossref] [PubMed]
- Ver Heul A, Planer J, Kau AL. The Human Microbiota and Asthma. Clin Rev Allergy Immunol 2019;57:350-63. [Crossref] [PubMed]
- Vestergaard DV, Holst GJ, Basinas I, et al. Pig Farmers' Homes Harbor More Diverse Airborne Bacterial Communities Than Pig Stables or Suburban Homes. Front Microbiol 2018;9:870. [Crossref] [PubMed]
- Stokholm J, Blaser MJ, Thorsen J, et al. Maturation of the gut microbiome and risk of asthma in childhood. Nat Commun 2018;9:141. [Crossref] [PubMed]
- Buendía E, Zakzuk J, San-Juan-Vergara H, et al. Gut microbiota components are associated with fixed airway obstruction in asthmatic patients living in the tropics. Sci Rep 2018;8:9582. [Crossref] [PubMed]
- Melli LC, do Carmo-Rodrigues MS, Araújo-Filho HB, et al. Intestinal microbiota and allergic diseases: A systematic review. Allergol Immunopathol (Madr) 2016;44:177-88. [Crossref] [PubMed]
- Hua X, Goedert JJ, Pu A, et al. Allergy associations with the adult fecal microbiota: Analysis of the American Gut Project. EBioMedicine 2016;3:172-9. [Crossref] [PubMed]
- West CE, Renz H, Jenmalm MC, et al. The gut microbiota and inflammatory noncommunicable diseases: associations and potentials for gut microbiota therapies. J Allergy Clin Immunol 2015;135:3-13; quiz 14. [Crossref] [PubMed]
- Fujimura KE, Lynch SV. Microbiota in allergy and asthma and the emerging relationship with the gut microbiome. Cell Host Microbe 2015;17:592-602. [Crossref] [PubMed]
- Goodrich JK, Waters JL, Poole AC, et al. Human genetics shape the gut microbiome. Cell 2014;159:789-99. [Crossref] [PubMed]
- Aguirre de Cárcer D, Cuív PO, Wang T, et al. Numerical ecology validates a biogeographical distribution and gender-based effect on mucosa-associated bacteria along the human colon. ISME J 2011;5:801-9. [Crossref] [PubMed]
- Arrieta MC, Arévalo A, Stiemsma L, et al. Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. J Allergy Clin Immunol 2018;142:424-434.e10. [Crossref] [PubMed]
- Li XL, Zhang B, Sun MJ, et al. Mechanism of gut microbiota and Axl/SOCS3 in experimental autoimmune encephalomyelitis. Biosci Rep 2019;39:BSR20190228 [Crossref] [PubMed]
- Schuijs MJ, Willart MA, Vergote K, et al. Farm dust and endotoxin protect against allergy through A20 induction in lung epithelial cells. Science 2015;349:1106-10. [Crossref] [PubMed]
- Gerhold K, Avagyan A, Seib C, et al. Prenatal initiation of endotoxin airway exposure prevents subsequent allergen-induced sensitization and airway inflammation in mice. J Allergy Clin Immunol 2006;118:666-73. [Crossref] [PubMed]
- Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol 2013;14:676-84. [Crossref] [PubMed]
- Chakraborti CK. New-found link between microbiota and obesity. World J Gastrointest Pathophysiol 2015;6:110-9. [Crossref] [PubMed]
- Choi IW. TNF-alpha induces the late-phase airway hyperresponsiveness and airway inflammation through cytosolic phospholipase A(2) activation. J Allergy Clin Immunol 2005;116:537-43. [Crossref] [PubMed]
- Peters MC, McGrath KW, Hawkins GA, et al. Plasma interleukin-6 concentrations, metabolic dysfunction, and asthma severity: a cross-sectional analysis of two cohorts. Lancet Respir Med 2016;4:574-84. [Crossref] [PubMed]
- Yalcin AD, Bisgin A, Gorczynski RM. IL-8, IL-10, TGF-β, and GCSF levels were increased in severe persistent allergic asthma patients with the anti-IgE treatment. Mediators Inflamm 2012;2012:720976 [Crossref] [PubMed]
- Silvestri M, Bontempelli M, Giacomelli M, et al. High serum levels of tumour necrosis factor-alpha and interleukin-8 in severe asthma: markers of systemic inflammation? Clin Exp Allergy 2006;36:1373-81. [Crossref] [PubMed]
- Ordoñez CL, Shaughnessy TE, Matthay MA, et al. Increased neutrophil numbers and IL-8 levels in airway secretions in acute severe asthma: Clinical and biologic significance. Am J Respir Crit Care Med 2000;161:1185-90. [Crossref] [PubMed]
- Nakagome K, Matsushita S, Nagata M. Neutrophilic inflammation in severe asthma. Int Arch Allergy Immunol 2012;158:96-102. [Crossref] [PubMed]
- McLoughlin RM, Mills KH. Influence of gastrointestinal commensal bacteria on the immune responses that mediate allergy and asthma. J Allergy Clin Immunol 2011;127:1097-107; quiz 1108-9. [Crossref] [PubMed]
- Doganci A, Eigenbrod T, Krug N, et al. The IL-6R alpha chain controls lung CD4+CD25+ Treg development and function during allergic airway inflammation in vivo. J Clin Invest 2005;115:313-25. [Crossref] [PubMed]
- Ramakrishnan RK, Al Heialy S, Hamid Q. Role of IL-17 in asthma pathogenesis and its implications for the clinic. Expert Rev Respir Med 2019;13:1057-68. [Crossref] [PubMed]
- Bullens DM, Truyen E, Coteur L, et al. IL-17 mRNA in sputum of asthmatic patients: linking T cell driven inflammation and granulocytic influx? Respir Res 2006;7:135. [Crossref] [PubMed]
- Zeng X, Gao X, Peng Y, et al. Higher Risk of Stroke Is Correlated With Increased Opportunistic Pathogen Load and Reduced Levels of Butyrate-Producing Bacteria in the Gut. Front Cell Infect Microbiol 2019;9:4. [Crossref] [PubMed]
- Serena C, Ceperuelo-Mallafré V, Keiran N, et al. Elevated circulating levels of succinate in human obesity are linked to specific gut microbiota. ISME J 2018;12:1642-57. [Crossref] [PubMed]
- To M, Hitani A, Kono Y, et al. Obesity-associated severe asthma in an adult Japanese population. Respir Investig 2018;56:440-7. [Crossref] [PubMed]
- Rhee H, Love T, Harrington D. Blood Neutrophil Count is Associated with Body Mass Index in Adolescents with Asthma. JSM Allergy Asthma 2018; [PubMed]
- Tang M, Henderson RJ, Holbrook JT, et al. Does Obesity Increase Respiratory Tract Infections in Patients with Asthma? J Allergy Clin Immunol Pract 2019;7:954-961.e6. [Crossref] [PubMed]
- Davila I, Valero A, Entrenas LM, et al. Relationship between serum total IgE and disease severity in patients with allergic asthma in Spain. J Investig Allergol Clin Immunol 2015;25:120-7. [PubMed]
- Borish L, Chipps B, Deniz Y, et al. Total serum IgE levels in a large cohort of patients with severe or difficult-to-treat asthma. Ann Allergy Asthma Immunol 2005;95:247-53. [Crossref] [PubMed]
- Verhamme KMC, Lucet C, Van Meerhaeghe A, et al. Real-life effectiveness of omalizumab in difficult-to-treat versus severe asthma: a national cohort study in Belgium. ERJ Open Res
- Dannemiller KC, Gent JF, Leaderer BP, et al. Indoor microbial communities: Influence on asthma severity in atopic and nonatopic children. J Allergy Clin Immunol 2016;138:76-83.e1. [Crossref] [PubMed]
- Mubarak B, Afzal N, Javaid K, et al. Frequency of HLA DQβ1*0201 and DQβ1*0301 Alleles and Total Serum IgE in Patients with Bronchial Asthma: A Pilot Study from Pakistan. Iran J Allergy Asthma Immunol 2017;16:313-20. [PubMed]
- Sandeep T, Roopakala MS, Silvia CR, et al. Evaluation of serum immunoglobulin E levels in bronchial asthma. Lung India 2010;27:138-40. [Crossref] [PubMed]
- Sears MR, Burrows B, Flannery EM, et al. Relation between airway responsiveness and serum IgE in children with asthma and in apparently normal children. N Engl J Med 1991;325:1067-71. [Crossref] [PubMed]
- Yin K, He S, Zhou L. The clinical allergy disease. Beijing Science Press 2012:125.
- Luo X, Pan Z, Luo S, et al. Effects of ceftriaxone-induced intestinal dysbacteriosis on regulatory T cells validated by anaphylactic mice. Int Immunopharmacol 2018;60:221-7. [Crossref] [PubMed]
- Chiu CY, Chan YL, Tsai MH, et al. Cross-talk between airway and gut microbiome links to IgE responses to house dust mites in childhood airway allergies. Sci Rep 2020;10:13449. [Crossref] [PubMed]
- Chung KF. Airway microbial dysbiosis in asthmatic patients: A target for prevention and treatment? J Allergy Clin Immunol 2017;139:1071-81. [Crossref] [PubMed]
- West CE, Rydén P, Lundin D, et al. Gut microbiome and innate immune response patterns in IgE-associated eczema. Clin Exp Allergy 2015;45:1419-29. [Crossref] [PubMed]
- Abrahamsson TR, Jakobsson HE, Andersson AF, et al. Low diversity of the gut microbiota in infants with atopic eczema. J Allergy Clin Immunol 2012;129:434-40, 440.e1-2.
- Liu X, Tao J, Li J, et al. Dysbiosis of Fecal Microbiota in Allergic Rhinitis Patients. Am J Rhinol Allergy 2020;34:650-60. [Crossref] [PubMed]


