
Cytoplasmic expression of G protein-coupled estrogen receptor 1 correlates with poor postoperative prognosis in non-small cell lung cancer
IntroductionOther Section
Lung cancer is one of the most common cancers globally and is currently the leading cause of cancer-related death in both males and females (1). A growing body of evidence now indicates that lung cancer is becoming prominent as a gender-related disease (2,3). It is well known that both lung adenocarcinoma (LUAD) and driver mutations of epidermal growth factor receptor (EGFR) and anaplastic lymphoma kinase (ALK) occur more commonly in females than in males (4,5), and positive expression of programmed cell death ligand-1 (PD-L1), a critical predictive biomarker for the efficacy of immunotherapy, was recently demonstrated to be higher in males than in females (6,7). Additionally, several prospective studies have shown that hormone replacement therapy (HRT) increases the incidence and mortality of lung cancer (8-10). Together, these data suggest that estrogen or gender‑dependent signaling are, at least in part, involved in the initiation and progression of lung cancer.
Estrogens act mainly through binding and activating their cognate receptors, estrogen receptor α (ERα) and ERβ. In view of the higher incidence rate of LUAD and EGFR mutation in females, both ERα and ERβ as well as estrogen signaling have been extensively investigated over the past 2 decades (11-14). Several retrospective studies with large participant cohorts have consistently reported that, in contrast to breast cancer, the expression level of ERβ is higher than that of ERα in lung cancer (15,16), and that strong expression of ERβ is positively correlated with EGFR mutation and could predict a better prognosis for patients with LUAD harboring EGFR mutations (15,17). In addition, several preclinical studies have revealed that estrogen promotes the proliferation of LUAD cells through activation of ERβ in vivo and in vitro (11,16). However, estrogen receptor (ER) inhibitor fulvestrant has only shown limited clinical efficacy for NSCLC patients in phase II clinical trials (18).
The G protein-coupled estrogen receptor 1 (GPER1), formerly known as GPR30, is the third ER, which is a potential membrane ER that can trigger a rapid, non-genomic signaling upon binding E2, environmental estrogens, as well as the antagonists of ERα and ERβ, such as fulvestrant and tamoxifen (19,20). It was found to be highly expressed and displayed biological activity in multiple solid tumors, especially in those showing gender differences in incidence, including breast, endometrial, thyroid, and colon cancer (21-24). In addition, functional interactions between GPER1 and EGFR or its downstream effectors such as protein kinase B (AKT) and extracellular-regulated kinase 1/2 (ERK1/2) have been well established and are thought to be the main mechanism by which GPER1 facilitates tumor progression (23,25-27).
More recently, the expression of GPER1 was found to be enhanced in NSCLC compared to normal lung tissue (28), but it is not yet clear whether its enhanced expression is the cause or consequence of lung carcinogenesis, and the subcellular localization of GPER1 has remained controversial. Recent studies have shown that activation of GPER1 with E2 or fulvestrant, an antagonist of both ERα and ERβ, promotes LUAD cell proliferation in vivo and in vitro (29-31), but prognostic effects of GPER1 in lung cancer remained unknown. Besides the higher incidence of LUAD and EGFR mutations in female patients, the recent finding of cross-talk between GPER1 and EGFR signaling (29) motivated us to investigate the correlation between the expression of GPER1 and clinicopathologic factors, especially EGFR mutations, and to evaluate the prognostic significance of GPER1 in NSCLC. We present the following article in accordance with the REMARK reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-29/rc).
MethodsOther Section
This was a retrospective and observational study of 183 consecutive patients at Yan’an Affiliated Hospital of Kunming Medical University who underwent surgical resection of tumors and were diagnosed with NSCLC, including132 cases of LUAD with identified EGFR mutations status and 51 squamous cell carcinoma (SCC) between June 2013 and June 2021. The study conformed to the Declaration of Helsinki (as revised in 2013) and was approved by the Institutional Ethics Committee of Yan’an Affiliated Hospital of Kunming Medical University (No. 2017-014-01). Specimens were stored according to protocols approved by the Institutional Review Board of Yan’an Affiliated Hospital of Kunming Medical University, and informed consent to use biopsy tissues for sample analyses was provided by all patients. All diagnoses were histologically proven and the pathological stage was adopted for the surgical cases according to the tumor-node-metastasis (TNM) classification revised in 2015 by the International Association for the Study of Lung Cancer (IASLC). None of the participants had been treated with EGFR–tyrosine kinase inhibitors (TKIs) or ALK inhibitors prior to lung tumor relapse.
Immunohistochemical staining and evaluation
The immunohistochemical (IHC) staining for GPER1 and ERβ was performed according to previously described methods (28). Briefly, the sections from paraffin-embedded lung carcinoma tissue were routinely prepared on glass slides and then deparaffinized. The sections were placed in 3% H2O2 for 10 min to quench the endogenous peroxidase. For epitope retrieval, they were heated for 30 min in 0.1 mol/L sodium citrate buffer (pH 6.0) in a water bath at 95–100 ℃. Then, the sections were incubated in normal goat serum for 20 min to reduce non-specific antibody binding. The primary antibody reaction employed the polyclonal rabbit antibody against ERβ (1:200; Proteintech, USA; code, 14007-1-AP) and GPER1 (1:200; Abcam, Cambridge, MA, USA; code, ab39742), confirmed to be specific for GPER1(32), for 90 min at room temperature. Thereafter, visualization reaction was performed using 3,3’-diaminobenzidine (DAB).
The IHC staining for GPER1 and ERβ was assessed using a defined scoring method (29) by 2 independent pathologists, who were blinded to the clinicopathologic data. Initially, a proportion score ranging from 1 to 4 was assigned according to the percentage of positive staining for tumor cells (1, 0–20%; 2, 21–50%; 3, 51–75%; and 4, 76–100%). Thereafter, 4 degrees of intensity score were also assigned according to the staining intensity (1, negative; 2, weak, 3, moderate; and 4, strong). The final value was obtained by multiplying the proportion and intensity scores, which ranged from 1 to 16 and was denoted as (−) ≤4, (+) >4 and ≤8, (++) >8 and ≤12, and (+++) >12 and ≤16. For statistical purposes, IHC scores of GPER1 were categorized into the weakly positive group (W group) when the score was 0–8 and the strongly positive group (S group) when the score was 9–16.
Detection of driver mutation
We detected EGFR mutations using a commercially available next generation sequencing (NGS) platform (majority in 3D Medicine Inc, Shanghai, China), which was self-funded by patients.
Statistical analysis
We compared 2 groups using the χ2 test, and multivariate models were constructed using logistic regression including the confounding factors with a P value <0.15 in univariate analysis. The Kaplan–Meier method was used to estimate the probability of recurrence-free survival (RFS) and overall survival (OS), and differences were analyzed by the log-rank test. The endpoint for RFS was the first documented day of recurrence of the disease. A multivariate analysis was performed according to the Cox proportional hazards model. The statistical difference was considered significant if the P value was less than 0.05. The data were analyzed using the software SPSS version 25.0 (IBM Corp., Chicago, IL, USA).
ResultsOther Section
Clinicopathological characteristics of 183 NSCLC patients
A total of 183 patients with pathologically confirmed primary NSCLC were enrolled from the Department of Thoracic Surgery (Yan’an Affiliated Hospital of Kunming Medical University) from June 2013 to June 2021. The median age of the183 participants at diagnosis was 60 years (31–87 years). Among all participants, 112 (61.2%) were <65 years old, 101 (55.2%) were female, 85 (46.4%) had a history of smoking, 132 (72.1%) presented with adenocarcinomas, and 51 (27.9%) presented with SCC (33). The pathological stage was I–II in 109 (59.6%) and III–IV in 74 participants (40.4%). Of the 183 tumors, 38 (20.8%) were poorly differentiated,145 (79.2%) were moderate to well differentiated, and local lymph node metastasis occurred in 76 (41.5%). Additionally, of the 132 LUAD participants, 52 (39.4%) harbored EGFR mutations, including 22 (16.7%) with exon 19 deletion and 30 (22.7%) with L858R point mutation.
Expression of GPER1 and its correlation with clinicopathological factors in NSCLC
The expression of GPER1 was found mainly in the nuclei and sometimes in the cytoplasm of carcinoma cells, and interestingly, all the samples expressing cGPER1 were also positive for nGPER1. Thus, positive expression patterns for GPER1 were categorized into 2 main classes: nGPER1 expression and concurrent n/cGPER1. Representative staining patterns of GPER1 are shown in Figure 1.
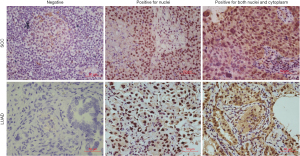
Among the 183 patients with NSCLC, a total of 153 patients (83.6%) had GPER1-positive NSCLC, including 64 with positive nGPER1 expression and 89 with positive n/cGPER1 expression. Of the 132 LUAD participants, 109 (82.6%) were positive for GPER1, including 47 (35.6%) with positive nGPER1 expression and 62 (47.0%) with positive n/cGPER1 expression. Of the 51 lung SCC, 44 (86.3%) were positive for GPER1, including 17 (33.3%) with positive nGPER1 expression and 27 (52.9%) with positive n/cGPER1 expression. However, positivity rates of both nGPER1 and n/cGPER1 expression did not show significant differences between LUAD and SCC (P=0.773, P=0.469, respectively).
In LUAD, univariate analysis revealed that the nGPER1 expression was significantly associated with EGFR mutations and never smokers, whereas the n/cGPER1 expression was significantly correlated with wt-EGFR, a history of smoking, stage III–VI, and lymph node metastasis. A multivariate analysis showed that nGPER1 expression was independently associated only with EGFR mutations [odds ratio (OR) =4.343; 95% confidence interval (CI): 2.035–9.270; P<0.001], and that n/cGPER1 expression was independently associated with EGFR mutations (OR =0.228; 95% CI: 0.104–0.500; P<0.001) and lymph node metastasis (OR =2.380; 95% CI: 1.096–5.168; P=0.028). Neither nGPER1 nor n/cGPER1 expression was associated with gender, age, and degree of differentiation (Table 1).
Table 1
| Variable | N | nGPER1 (LUAD), n (%) | P value | n/cGPER1 (LUAD), n (%) | P value | ERβ (LUAD), n (%) | P value |
|---|---|---|---|---|---|---|---|
| Age, years | 0.510 | 0.287 | 0.260 | ||||
| <65 | 85 | 32 (37.65) | 37 (43.53) | 33 (38.82) | |||
| ≥65 | 47 | 15 (31.91) | 25 (53.19) | 21 (44.68) | |||
| Gender | 0.272 | 0.249 | 0.686 | ||||
| Male | 59 | 22 (37.29) | 31 (52.54) | 23 (38.98) | |||
| Female | 73 | 29 (39.73) | 31 (42.47) | 31 (42.67) | |||
| Smoking history | 0.030 | 0.047 | 0.596 | ||||
| Smoker | 50 | 12 (24.00) | 29 (58.00) | 19 (38.00) | |||
| Never smoker | 82 | 35 (42.68) | 33 (40.24) | 35 (42.68) | |||
| Differentiation | 0.466 | 0.97 | 0.759 | ||||
| Low | 30 | 9 (30.00) | 14 (46.67) | 13 (43.33) | |||
| Middle & high | 102 | 38 (37.25) | 48 (47.06) | 41 (20.59) | |||
| EGFR mutation | <0.001 | <0.001 | 0.038 | ||||
| No | 80 | 18 (22.50) | 49 (61.25) | 27 (33.75) | |||
| Yes | 52 | 29 (55.77) | 13 (25.00) | 27 (51.92) | |||
| Stage | 0.197 | 0.007 | 0.815 | ||||
| Stage 1/2 | 84 | 34 (40.48) | 33 (39.29) | 35 (41.67) | |||
| Stage 3/4 | 48 | 13 (27.08) | 29 (60.42) | 19 (39.58) | |||
| Lymph node metastasis | 0.243 | 0.007 | 0.815 | ||||
| Negative | 84 | 14 (16.67) | 32 (38.10) | 35 (41.67) | |||
| Positive | 48 | 33 (68.75) | 30 (62.50) | 19 (39.58) |
GPER1, G protein-coupled estrogen receptor 1; ERβ, estrogen receptor β; nGPER1, positive for nuclear expression of GPER1; n/cGPER1, positive expression of GPER1 both in nuclei and cytoplasm; LUAD, lung adenocarcinoma; EGFR, epidermal growth factor receptor.
In SCC, both the nGPER1 and n/cGPER1 expression were not significantly associated with gender, age, smoking history, lymph node metastasis, tumor stage, and degree of differentiation (Table 2).
Table 2
| Variable | N | nGPER1 (SCC), n (%) | P value | n/cGPER1 (SCC), n (%) | P value | ERβ (SCC), n (%) | P value |
|---|---|---|---|---|---|---|---|
| Age, years | 0.552 | 0.467 | 1.000 | ||||
| <65 | 27 | 10 (37.04) | 13 (48.15) | 4 (14.81) | |||
| ≥65 | 24 | 7 (29.17) | 14 (58.33) | 5 (20.83) | |||
| Gender | 0.051 | 0.363 | 0.294 | ||||
| Male | 42 | 17 (40.48) | 21 (50.00) | 9 (21.43) | |||
| Female | 9 | 0 (0.00) | 6 (66.67) | 0 (0.00) | |||
| Smoking history | 0.286 | 0.374 | 0.798 | ||||
| Smoker | 35 | 10 (28.47) | 20 (57.14) | 7 (0.20) | |||
| Never smoker | 16 | 7 (43.75) | 7 (43.75) | 2 (0.13) | |||
| Differentiation | 0.785 | 0.856 | 1.000 | ||||
| Low | 8 | 3 (37.50) | 4 (50.00) | 1 (12.50) | |||
| Middle & high | 43 | 14 (32.56) | 23 (53.49) | 8 (18.60) | |||
| Lymph node metastasis | 0.164 | 0.22 | 0.072 | ||||
| Negative | 23 | 7 (30.43) | 17 (73.91) | 7 (30.43) | |||
| Positive | 28 | 10 (35.71) | 10 (35.71) | 2 (7.14) |
GPER1, G protein-coupled estrogen receptor 1; ERβ, estrogen receptor β; SCC, squamous cell carcinoma; nGPER1, positive for nuclear expression of GPER1; n/cGPER1, positive expression of GPER1 both in nuclei and cytoplasm.
Expression of ERβ and its correlation with clinicopathological factors in NSCLC
The expression of ERβ was found mainly in the cytoplasm of cancer cells, and its positivity rate was significantly higher in LUAD than in SCC (40.9% vs. 17.6%, P=0.003). Representative staining of ERβ is shown in Figure 2.

In LUAD, univariate analysis showed that the expression of ERβ was positively correlated with EGFR mutations and nGPER1 expression, but negatively with n/cGPER1 expression. A multivariate analysis revealed that the expression of ERβ was independently associated only with nGPER1 expression (OR =6.333; 95% CI: 2.092–19.170; P=0.001).
In SCC, univariate analysis showed that ERβ expression was not significantly associated with any clinicopathological factors; however, a multivariate analysis suggested that the expression of ERβ was independently associated with advanced stage of tumor (OR =0.176; 95% CI: 0.032–0.953; P=0.044).
EGFR mutations in lung adenocarcinoma
The incidence of EGFR mutations in our cohort was 39.4% (52/132), and its distribution in stage I, II, III and IV was 46.6%, 45.5%, 22.2% and 33.3%, respectively, but there was no significant difference in frequencies of EGFR mutation between different tumor stages (P=0.145).
A total of 65 patients (25 EGFR mutant and 40 EGFR wildtype) treated with palliative chemotherapy in 132 LUAD, and all of them eventually experienced progression. The overall response rate (ORR) to first-line chemotherapy was higher in patients with EGFR mutations than those with EGFR wildtype (60.0% vs. 27.5%, P=0.009), and the median progression-free survival (mPFS) was longer in patients with EGFR mutations than those with EGFR wildtype (128 vs. 68 days, P=0.001).
Influence of expression of GPER1 on RFS
To evaluate the prognostic effect of expression of GPER1, we compared the RFS of 146 participants, including 104 LUAD and 42 SCC, who underwent complete surgical resection of their tumors.
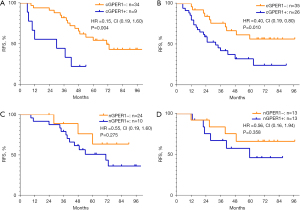
In 104 LUAD patients, 55 (52.9%) had relapsed. The EGFR mutations did not influence the RFS (P=0.455). However, a positive expression of n/cGPER1 type was significantly associated with poor prognosis; the RFS was significantly worse in patients with n/cGPER1 expression than in those without cytoplasmic expression of GPER1, including nGPER1 and negative GPER1 expression [hazard ratio (HR) =2.73, 95% CI: 1.55–4.81, P=0.001]. Further, the survival data were compared among n/cGPER1, nGPER1, and negative GPER1 groups, and the RFS was significantly worse in the n/cGPER1 group than that in the other 2 groups (HR =4.82 for n/cGPER1 vs. negative GPER1, 95% CI: 2.03–11.43, P<0.001; HR =2.03 for positive n/cGPER1 vs. nGPER1, 95% CI: 1.10–3.72, P=0.023), but there was only a marginal difference in RFS for the nGPER1 group versus negative group (HR =2.38, 95% CI: 0.99–5.71, P=0.052). Further, the participants were stratified by their EGFR mutated status because a strong correlation between the subcellular localization of GPER1 and EGFR mutations was observed, as shown in Table 1. The n/cGPER1 expression was significantly associated with decreased RFS in both EGFR mutation (P=0.004; Figure 3A) and wt-EGFR group (P=0.01; Figure 3B), but nGPER1 expression was not in these 2 groups (P=0.275 for EGFR mutation, Figure 3C; P=0.358 for wt-EGFR, Figure 3D). The effects of various clinicopathologic factors on RFS in LUAD patients were evaluated by univariate and multivariate analysis. As a result, n/cGPER1 expression (HR =2.73, 95% CI: 1.55–4.81, P=0.001), advanced stage (HR =3.35 for stage II vs. stage I, 95% CI: 1.34–8.35, P=0.009; HR 2.99 for stage III vs. stage I, 95% CI: 1.34–6.69, P=0.007) and lymph node metastasis (HR =2.93, 95% CI: 1.31–6.55, P=0.009) were independently correlated with poor RFS in patients with LUAD.
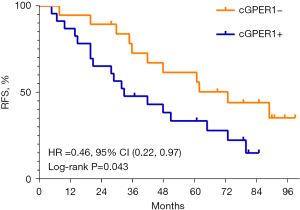
Among 42 SCC patients, 29 (69.0%) had relapsed. The mRFS was notably shorter in the group with n/cGPER1 expression than in the group without cytoplasmic expression (P=0.043; Figure 4). Additionally, the mRFS was also notably shorter in tumors with poor differentiation, lymph node metastasis, and advanced stage than their counterparts (P=0.010, P<0.001, P<0.001, respectively). A multivariate analysis showed that only n/cGPER1 expression (HR =3.15, 95% CI: 1.40–7.12, P=0.006) and tumor stage (HR =2.44 for stage II vs. stage I, 95% CI: 0.68–8.73, P=0.17; HR =15.99 for stage III vs. stage I, 95% CI: 4.16–61.54, P<0.001) were independently correlated with poor RFS in patients with SCC. However, we could not evaluate the effect of nGPER1 expression on RFS in SCC due to the small size of our cohort.
Influence of expression of GPER1 on OS
Among the 132 LUAD patients, there were 58 (43.9%) deaths. The median overall survival (mOS) was markedly shorter in the group with n/cGPER1 expression than in the group without cytoplasmic expression (P<0.001; Figure 5A). In addition, the mOS for the entire cohort was also affected by the EGFR mutations(P=0.043), advanced stage (P<0.001), lymph node metastasis (P<0.001). In multivariate analysis, the OS remained affected by n/cGPER1 expression (HR =3.617, 95% CI: 1.989–6.576, P<0.001), advanced stage (HR =2.516, 95% CI: 1.059–5.977, P=0.037) and lymph node metastasis (HR =4.188, 95% CI: 1.735–10.111, P=0.001).
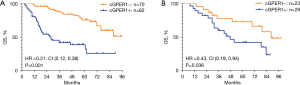
Among the 51 SCC patients, there were 27 (52.9%) deaths. The expression of n/cGPER1 significantly decreased the mOS of the entire cohort (P=0.036; Figure 5B). Besides, the OS was also affected by the low differentiation (P=0.043), advanced stage (P<0.001), lymph node metastasis (P<0.001). However, in multivariate analysis, the OS was not affected by n/cGPER1 expression (P=0.122), except for the advanced stage (HR =6.169, 95% CI: 1.139–33.423, P=0.035) and lymph node metastasis (HR =4.136, 95% CI: 1.090–15.697, P=0.037).
DiscussionOther Section
Estrogen has long been thought to promote the initiation and development of lung cancer, whereas anti-estrogen therapy based on inhibiting ERβ signaling has shown limited clinical efficacy (18). In this study, we analyzed the correlations between the expression of GPER1 and various clinicopathological factors including EGFR mutations and ERβ expression, and further evaluated its prognostic significance in postoperative NSCLC patients.
We found a unique expression profile of GPER1 in NSCLC for the first time: GPER1 expression was concurrently present in nuclei and cytoplasm, and it appeared that cGPER1 expression was based on the expression of nGPER1; a similar observation was also made in the endometrium using the same GPER1 antibody as used in our study (32). However, our results were inconsistent with the previous study, where GPER1 expression was found mainly in the cytoplasm and sometimes in the nuclei of lung cancer cells (29,31). More recently, it has been demonstrated that GPER1 is a glycosylated protein receptor (34). The N-terminal glycosylation can influence its structure, activity, and subcellular localization, thus rendering its subcellular localization and function more complex (34,35). In breast cancer cells, only the cytoplasm or membrane expression of GPER1 can transactivate EGFR signaling (26,36). However, disrupting N-glycosylation trigger the translocalization of GPER1 from cytoplasm to nucleus, where it was unable to activate mitogen-activated protein kinase (MAPK) signaling, a downstream effector of EGFR signaling, but could enhance cellular proliferation and migration by binding to the promoters of its target genes c-FOS and connective tissue growth factor (CTGF), respectively (37). Thus, different subcellular localization of GPER1 could be caused by its glycosylation status.
Several studies have shown that the nuclear expression of ERβ is positively correlated with EGFR mutations in LUAD (4,15,17). In the present study, we found that both ERβ and nGPER1 expression occurred more frequently in EGFR-mutated LUAD, whereas n/cGPER1 expression occurred more frequently in wt-EGFR. Conversely, a functional interaction between GPER1 and EGFR as well as ERs has been established in several solid tumors (23,36). In breast cancer cells, for instance, GPER1 could translocate from nuclei to cytoplasm and membrane after long-term inhibition of ERα/β signaling, which in turn transactivates EGFR signaling, resulting in cell proliferation, migration, and even resistance to endocrine therapy (23,38). Thus, the activity of EGFR and ERβ signaling could affect the subcellular localization of GPER1. In addition, a reasonable interpretation of such correlation between subcellular localizations of GPER1 and mutation status of EGFR may be that, when EGFR is at activating mutation status or aberrantly active, GPER1 mainly localizes in nuclei; however, when EGFR is at wild-type status or inactive, GPER1 can translocate from nuclei to cytoplasm to functionally interact with EGFR, thereby complementarily enhancing EGFR signaling. Although EGFR-TKIs have largely improved outcomes and quality of life of NSCLC patients with EGFR mutations, resistance to these drugs eventually emerged. Besides the secondary mutation of EGFR, amplification of EGFR and c-MET, activation of EGFR downstream signaling, mainly including MAPK and PI3K/AKT pathways, have been demonstrated to play a critical role in such acquired resistance (39,40). Thus, based on what discussed above, this finding may provide an opportunity for investigating the potential mechanism underlying EGFR mutations; further, targeting GPER1 could be a strategy for overcoming EGFR-TKIs resistance in NSCLC in the future.
Results from our study showed that the positive expression of ERβ was higher in LUAD than in SCC, and also higher in EGFR-mutated LUAD than in wt-EGFR LUAD, which were in line with previous reports (14). Additionally, our present work for the first time showed that the expression of nGPER1 was higher in EGFR mutations than in wt-EGFR LUAD, and there was a trend toward a higher expression level of nGPER1 in LUAD than in SCC harboring a lower frequency of EGFR mutations, though the difference was not significant. Further, the expression of ERβ was positively correlated with nGPER1 expression, but not with n/cGPER1 expression.
It has been previously reported that GPER1 is a potential risk factor in promoting distant metastasis and could enhance malignancy of multiple tumors, including breast, ovary, and cervical cancer (41-43). However, data regarding the role and impact of GPER1 on the progression of NSCLC is very limited so far, only several preclinical studies being reported. GPER1 could promote NSCLC progression through activation of MAPK, PI3K/AKT and NOTCH1 signaling pathway in NSCLC (29,30). Whereas another study reported that activation of GPER1 inhibited the migration of NSCLC cells via IKK-β/NF-κB signals (44). These data were conflicted on the role of GPER1 in NSCLC progression, the potential mechanism for which is unknown. In this work, our results showed that cytoplasmic and nuclear expression of GPER1 could result in different prognosis for patients with NSCLC, combining with previous finding in breast cancer cells that glycosylated form of GPER1 was localized in cytoplasm while non-glycosylated form localized in nuclei, and that they could exert different roles. Therefore, these conflicting results could be caused by the different glycosylated status of GPER1. In the present research, we found that only the n/cGPER1 expression, but not the nGPER1 expression, was significantly associated with the advanced stage of tumor and lymph node metastasis, which was consistent with the previous study in LUAD (29). In 2 previous studies, GPER1 IHC patterns were divided into nuclear and cytoplasmic expressions, and their correlation with clinicopathological factors in NSCLC were investigated separately (29,31). However, in most cases, the expression of nGPER1 and cGPER1 occurred concurrently in the same patient. Thus, in order to evaluate prognostic effect of GPER1, we categorized GPER1 IHC patterns into 3 subtypes: negative, nGPER1, and n/cGPER1. Herein, we evaluated for the first time the prognostic significance of GPER1 in NSCLC, and found n/cGPER1, but not nGPER1 expression, was significantly associated with poor RFS and OS in NSCLC. Even after stratifying the LUAD patients by EGFR mutation status, n/cGPER1 expression was still linked to a shorter RFS in both the EGFR-mutated and wt-EGFR groups. However, we could not evaluate the impact of nGPER1 on RFS after stratifying the LUAD patients by EGFR-mutated status, due to the small size of our cohort.
In conclusion, GPER1 is aberrantly highly expressed in both LUAD and SCC. The nGPER1 expression occurs more frequently in EGFR-mutated LUAD, while n/cGPER1 expression occurs more frequently in wt-EGFR LUAD. The n/cGPER1 type predicts a worse RFS and OS in NSCLC, which is a potential risk factor for prognosis of NSCLC patients.
This work will facilitate a better understanding of estrogen signaling in the development of NSCLC, and GPER1 can be considered as a potential target or biomarker for treatment of NSCLC. Clinical studies with large simple sizes and preclinical research are needed to clarify the role of GPER1 in lung cancer, especially in the interaction with EGFR signaling pathway, which may provide a new strategy to overcome EGFR-TKIs resistance in the future.
AcknowledgmentsOther Section
Funding: This study was funded by the Key Project of Applied Basic Research of Yunnan Province (No. 2018FA044) and supported by the Key Laboratory of Tumor Immunological Prevention and Treatment of Yunnan Province (No. 2017DG004).
FootnoteOther Section
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-29/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-29/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-29/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study conformed to the Declaration of Helsinki (as revised in 2013) and was approved by the Institutional Ethics Committee of Yan’an Affiliated Hospital of Kunming Medical University (No. 2017-014-01). Informed consent to use biopsy tissues for sample analyses was provided by all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
ReferencesOther Section
- Bade BC, Dela Cruz CS. Lung Cancer 2020: Epidemiology, Etiology, and Prevention. Clin Chest Med 2020;41:1-24. [Crossref] [PubMed]
- Wen S, Dai L, Wang L, et al. Genomic Signature of Driver Genes Identified by Target Next-Generation Sequencing in Chinese Non-Small Cell Lung Cancer. Oncologist 2019;24:e1070-81. [Crossref] [PubMed]
- Smida T, Bruno TC, Stabile LP. Influence of Estrogen on the NSCLC Microenvironment: A Comprehensive Picture and Clinical Implications. Front Oncol 2020;10:137. [Crossref] [PubMed]
- Kawaguchi T, Koh Y, Ando M, et al. Prospective Analysis of Oncogenic Driver Mutations and Environmental Factors: Japan Molecular Epidemiology for Lung Cancer Study. J Clin Oncol 2016;34:2247-57. [Crossref] [PubMed]
- Haupt S, Caramia F, Klein SL, et al. Sex disparities matter in cancer development and therapy. Nat Rev Cancer 2021;21:393-407. [Crossref] [PubMed]
- Lee SE, Kim YJ, Sung M, et al. Association with PD-L1 Expression and Clinicopathological Features in 1000 Lung Cancers: A Large Single-Institution Study of Surgically Resected Lung Cancers with a High Prevalence of EGFR Mutation. Int J Mol Sci 2019;20:4794. [Crossref] [PubMed]
- Wen Y, Chen Y, Duan X, et al. The clinicopathological and prognostic value of PD-L1 in urothelial carcinoma: a meta-analysis. Clin Exp Med 2019;19:407-16. [Crossref] [PubMed]
- Chlebowski RT, Schwartz AG, Wakelee H, et al. Oestrogen plus progestin and lung cancer in postmenopausal women (Women's Health Initiative trial): a post-hoc analysis of a randomised controlled trial. Lancet 2009;374:1243-51. [Crossref] [PubMed]
- Baik CS, Eaton KD. Estrogen signaling in lung cancer: an opportunity for novel therapy. Cancers (Basel) 2012;4:969-88. [Crossref] [PubMed]
- Ganti AK, Sahmoun AE, Panwalkar AW, et al. Hormone replacement therapy is associated with decreased survival in women with lung cancer. J Clin Oncol 2006;24:59-63. [Crossref] [PubMed]
- Garon EB, Pietras RJ, Finn RS, et al. Antiestrogen fulvestrant enhances the antiproliferative effects of epidermal growth factor receptor inhibitors in human non-small-cell lung cancer. J Thorac Oncol 2013;8:270-8. [Crossref] [PubMed]
- Toh CK, Ahmad B, Soong R, et al. Correlation between epidermal growth factor receptor mutations and expression of female hormone receptors in East-Asian lung adenocarcinomas. J Thorac Oncol 2010;5:17-22. [Crossref] [PubMed]
- Márquez-Garbán DC, Chen HW, Fishbein MC, et al. Estrogen receptor signaling pathways in human non-small cell lung cancer. Steroids 2007;72:135-43. [Crossref] [PubMed]
- Raso MG, Behrens C, Herynk MH, et al. Immunohistochemical expression of estrogen and progesterone receptors identifies a subset of NSCLCs and correlates with EGFR mutation. Clin Cancer Res 2009;15:5359-68. [Crossref] [PubMed]
- Nose N, Sugio K, Oyama T, et al. Association between estrogen receptor-beta expression and epidermal growth factor receptor mutation in the postoperative prognosis of adenocarcinoma of the lung. J Clin Oncol 2009;27:411-7. [Crossref] [PubMed]
- Fu S, Liu C, Huang Q, et al. Estrogen receptor β1 activation accelerates resistance to epidermal growth factor receptor-tyrosine kinase inhibitors in non-small cell lung cancer. Oncol Rep 2018;39:1313-21. [Crossref] [PubMed]
- Nose N, Uramoto H, Iwata T, et al. Expression of estrogen receptor beta predicts a clinical response and longer progression-free survival after treatment with EGFR-TKI for adenocarcinoma of the lung. Lung Cancer 2011;71:350-5. [Crossref] [PubMed]
- Garon EB, Siegfried JM, Stabile LP, et al. Randomized phase II study of fulvestrant and erlotinib compared with erlotinib alone in patients with advanced or metastatic non-small cell lung cancer. Lung Cancer 2018;123:91-8. [Crossref] [PubMed]
- Luo J, Liu D. Does GPER Really Function as a G Protein-Coupled Estrogen Receptor in vivo? Front Endocrinol (Lausanne) 2020;11:148. [Crossref] [PubMed]
- Barton M, Filardo EJ, Lolait SJ, et al. Twenty years of the G protein-coupled estrogen receptor GPER: Historical and personal perspectives. J Steroid Biochem Mol Biol 2018;176:4-15. [Crossref] [PubMed]
- Hernández-Silva CD, Villegas-Pineda JC, Pereira-Suárez AL. Expression and Role of the G Protein-Coupled Estrogen Receptor (GPR30/GPER) in the Development and Immune Response in Female Reproductive Cancers. Front Endocrinol (Lausanne) 2020;11:544. [Crossref] [PubMed]
- Jacenik D, Beswick EJ, Krajewska WM, et al. G protein-coupled estrogen receptor in colon function, immune regulation and carcinogenesis. World J Gastroenterol 2019;25:4092-104. [Crossref] [PubMed]
- Xu S, Yu S, Dong D, et al. G Protein-Coupled Estrogen Receptor: A Potential Therapeutic Target in Cancer. Front Endocrinol (Lausanne) 2019;10:725. [Crossref] [PubMed]
- Hsu LH, Chu NM, Lin YF, et al. G-Protein Coupled Estrogen Receptor in Breast Cancer. Int J Mol Sci 2019;20:306. [Crossref] [PubMed]
- Gonzalez de Valdivia E, Broselid S, Kahn R, et al. G protein-coupled estrogen receptor 1 (GPER1)/GPR30 increases ERK1/2 activity through PDZ motif-dependent and -independent mechanisms. J Biol Chem 2017;292:9932-43. [Crossref] [PubMed]
- Mo Z, Liu M, Yang F, et al. GPR30 as an initiator of tamoxifen resistance in hormone-dependent breast cancer. Breast Cancer Res 2013;15:R114. [Crossref] [PubMed]
- Fujiwara S, Terai Y, Kawaguchi H, et al. GPR30 regulates the EGFR-Akt cascade and predicts lower survival in patients with ovarian cancer. J Ovarian Res 2012;5:35. [Crossref] [PubMed]
- Jala VR, Radde BN, Haribabu B, et al. Enhanced expression of G-protein coupled estrogen receptor (GPER/GPR30) in lung cancer. BMC Cancer 2012;12:624. [Crossref] [PubMed]
- Liu C, Liao Y, Fan S, et al. G protein-coupled estrogen receptor (GPER) mediates NSCLC progression induced by 17β-estradiol (E2) and selective agonist G1. Med Oncol 2015;32:104. [Crossref] [PubMed]
- Shen Y, Li C, Zhou L, et al. G protein-coupled oestrogen receptor promotes cell growth of non-small cell lung cancer cells via YAP1/QKI/circNOTCH1/m6A methylated NOTCH1 signalling. J Cell Mol Med 2021;25:284-96. [Crossref] [PubMed]
- Liu C, Liao Y, Fan S, et al. G-Protein-Coupled Estrogen Receptor Antagonist G15 Decreases Estrogen-Induced Development of Non-Small Cell Lung Cancer. Oncol Res 2019;27:283-92. [Crossref] [PubMed]
- Samartzis N, Samartzis EP, Noske A, et al. Expression of the G protein-coupled estrogen receptor (GPER) in endometriosis: a tissue microarray study. Reprod Biol Endocrinol 2012;10:30. [Crossref] [PubMed]
- Pietras RJ, Márquez DC, Chen HW, et al. Estrogen and growth factor receptor interactions in human breast and non-small cell lung cancer cells. Steroids 2005;70:372-81. [Crossref] [PubMed]
- Gonzalez de Valdivia E, Sandén C, Kahn R, et al. Human G protein-coupled receptor 30 is N-glycosylated and N-terminal domain asparagine 44 is required for receptor structure and activity. Biosci Rep 2019;39:BSR20182436. [Crossref] [PubMed]
- Cheng SB, Graeber CT, Quinn JA, et al. Retrograde transport of the transmembrane estrogen receptor, G-protein-coupled-receptor-30 (GPR30/GPER) from the plasma membrane towards the nucleus. Steroids 2011;76:892-6. [Crossref] [PubMed]
- Molina L, Figueroa CD, Bhoola KD, et al. GPER-1/GPR30 a novel estrogen receptor sited in the cell membrane: therapeutic coupling to breast cancer. Expert Opin Ther Targets 2017;21:755-66. [Crossref] [PubMed]
- Pupo M, Bodmer A, Berto M, et al. A genetic polymorphism repurposes the G-protein coupled and membrane-associated estrogen receptor GPER to a transcription factor-like molecule promoting paracrine signaling between stroma and breast carcinoma cells. Oncotarget 2017;8:46728-44. [Crossref] [PubMed]
- Ignatov A, Ignatov T, Roessner A, et al. Role of GPR30 in the mechanisms of tamoxifen resistance in breast cancer MCF-7 cells. Breast Cancer Res Treat 2010;123:87-96. [Crossref] [PubMed]
- Ku BM, Choi MK, Sun JM, et al. Acquired resistance to AZD9291 as an upfront treatment is dependent on ERK signaling in a preclinical model. PLoS One 2018;13:e0194730. [Crossref] [PubMed]
- Santoni-Rugiu E, Melchior LC, Urbanska EM, et al. Intrinsic resistance to EGFR-Tyrosine Kinase Inhibitors in EGFR-Mutant Non-Small Cell Lung Cancer: Differences and Similarities with Acquired Resistance. Cancers (Basel) 2019;11:923. [Crossref] [PubMed]
- He YY, Du GQ, Cai B, et al. Estrogenic transmembrane receptor of GPR30 mediates invasion and carcinogenesis by endometrial cancer cell line RL95-2. J Cancer Res Clin Oncol 2012;138:775-83. [Crossref] [PubMed]
- Ignatov T, Claus M, Nass N, et al. G-protein-coupled estrogen receptor GPER-1 expression in hormone receptor-positive breast cancer is associated with poor benefit of tamoxifen. Breast Cancer Res Treat 2019;174:121-7. [Crossref] [PubMed]
- Talia M, De Francesco EM, Rigiracciolo DC, et al. The G Protein-Coupled Estrogen Receptor (GPER) Expression Correlates with Pro-Metastatic Pathways in ER-Negative Breast Cancer: A Bioinformatics Analysis. Cells 2020;9:622. [Crossref] [PubMed]
- Zhu G, Huang Y, Wu C, et al. Activation of G-Protein-Coupled Estrogen Receptor Inhibits the Migration of Human Nonsmall Cell Lung Cancer Cells via IKK-β/NF-κB Signals. DNA Cell Biol 2016;35:434-42. [Crossref] [PubMed]


