
The role of miR-145-5p in esophageal squamous cell carcinoma tumor-associated macrophages and selection of immunochemotherapy
Introduction
Esophageal cancer ranks as the sixth most common malignancy worldwide (1). It is classified as esophageal squamous cell carcinoma (ESCC) or adenocarcinoma. ESCC is one of the leading causes of cancer-related deaths in Asia, especially in southern and central Asia. Despite recent advances in therapeutics against ESCC, the 5-year survival rate still ranges from 9% to 27.1% (1,2). Among the many different therapeutic approaches, the focus has now shifted to immunotherapy. Because of the high risk of metastatic recurrence in ESCC, where cell dissemination occurs early, the surgical treatment options are limited. Therefore, active immunotherapy may deserve a place among the different treatment modalities.
MicroRNAs (miRs) are noncoding RNAs with a length of approximately 22 nucleotides (3). Previous studies have confirmed that miRs can regulate the biological characteristics of tumor cells (4-6), can affect the progression of tumors, and are related to the prognosis of patients. Recent evidence indicates that miRs are associated with the immune microenvironment, which can help to predict the response of tumor patients to immunochemotherapy and may be a target of immunotherapy (7).
A recent study has shown that the levels of miR-145-5p expression are associated with disease-free survival (DFS) rates in breast cancer patients and could be a potential biomarker of the clinical response to neoadjuvant chemotherapy in triple-negative breast cancer (8). In fact, it has already been reported that miR-145-5p was related to the response to neoadjuvant chemotherapy in ESCC and was associated with clinicopathologic features and the recurrence of metastasis in ESCC patients (7,9-11). These findings suggest that miR-145-5p plays an important role in predicting the efficacy of chemoradiotherapy. It is well known that the immune microenvironment plays an important role in the immunochemotherapeutic response of tumor patients (12). However, the mechanism of miR-145-5p has not been elucidated and the potential application of miR-145-5p in ESCC immunochemotherapy remains unknown.
In our study, we investigate the relationship between miR-145-5p and the clinical features of patients with ESCC to explore the predictive role of miR-145-5p on prognosis in patients with ESCC, and we elaborate on the impact of miR-145-5p on immune infiltration aimed to better understand the cellular and molecular biology within the tumor microenvironment related to immunochemotherapy in ESCC and explore the possibility of miR-145-5p as a target for immunochemotherapy and a predictor of the prognosis for the treatment of immunotherapy. We present the following article in accordance with the REMARK reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-294/rc).
Methods
Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) data retrieval
HTSeq-FPKM and the Clinical Dataset for ESCC were downloaded from the TCGA database (https://cancergenome.nih.gov/). We retrieved the miR expression data of 95 ESCC tissues and 13 normal tissues, as well as the clinical materials of 95 patients with ESCC. Table 1 presents the clinicopathological parameters of patients with ESCC; the miR expression levels were standardized by log2n transformation. The GSE43732 (China, 2013) dataset and GSE13937 (USA, 2008) dataset were obtained from the GEO website (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi) using the key terms “esophageal squamous cell carcinoma” and “prognosis”. The GSE43732 dataset included miR expression data, as well as clinical and prognostic data for 119 patients with ESCC and corresponding normal tissues. All data for miR expression were standardized by 2n transformation. Table 2 presents the clinicopathological parameters of patients with ESCC. The GSE13937 dataset included miR expression data, as well as the clinical and prognostic data of 44 patients with ESCC and corresponding normal tissues. MiR expression profiles of frozen tissues from ESCC patients were all assessed by microarray.
Table 1
| Clinical characteristics | N | Relative expression of miR-145-5p | ||
|---|---|---|---|---|
| Mean ± SD | t/F value | P value | ||
| Tissue | −2.440 | 0.016* | ||
| Tumor | 95 | 11.25±1.46 | ||
| Normal | 13 | 12.30±1.43 | ||
| Gender | 1.412 | 0.161 | ||
| Male | 80 | 11.34±1.46 | ||
| Female | 15 | 10.76±1.42 | ||
| Age (years) | 1.651 | 0.102 | ||
| ≥60 | 39 | 11.54±1.44 | ||
| <60 | 56 | 11.04±1.45 | ||
| Alcohol | −1.744 | 0.085 | ||
| Yes | 68 | 11.10±1.44 | ||
| No | 25 | 11.70±1.49 | ||
| Location | F=0.088a | 0.916 | ||
| Proximal | 6 | 11.49±1.16 | ||
| Mid | 44 | 11.26±1.53 | ||
| Distal | 44 | 11.22±1.46 | ||
| G status | F=2.542a | 0.085 | ||
| G1 | 16 | 10.70±1.17 | ||
| G2 | 49 | 11.51±1.43 | ||
| G3 | 20 | 10.89±1.65 | ||
| M status | −2.511 | 0.039* | ||
| M0 | 84 | 11.19±1.46 | ||
| M1 | 5 | 11.96±0.59 | ||
| T status | F=0.922a | 0.433 | ||
| T1 | 7 | 11.20±0.92 | ||
| T2 | 34 | 11.09±1.45 | ||
| T3 | 49 | 11.26±1.51 | ||
| T4 | 5 | 12.25±1.57 | ||
| N status | −0.551 | 0.583 | ||
| N0 | 51 | 11.27±1.58 | ||
| N1–2 | 34 | 11.44±1.25 | ||
| Stage | −1.048 | 0.297 | ||
| I–II | 62 | 11.15±1.47 | ||
| III–IV | 32 | 11.48±1.43 | ||
a, one-way ANOVA test was utilized; *, comparison of miR-145-5p expression between or among each variable is statistical significance. ESCC, esophageal squamous cell carcinoma; TCGA, The Cancer Genome Atlas; SD, standard deviation; t, Student’s t-test; ANOVA, analysis of variance.
Table 2
| Clinical characteristics | N | Relative expression of miR-145-5p | ||
|---|---|---|---|---|
| Mean ± SD | t/F value | P value | ||
| Tissue | −7.730 | 0.000* | ||
| Tumor | 119 | 1.95±3.64 | ||
| Normal | 119 | 5.05±3.06 | ||
| Gender | −0.919 | 0.368 | ||
| Male | 98 | 1.71±2.58 | ||
| Female | 21 | 3.06±6.64 | ||
| Age (years) | −1.178 | 0.241 | ||
| ≥60 | 58 | 1.54±2.04 | ||
| <60 | 61 | 2.33±4.66 | ||
| Alcohol | −0.911 | 0.364 | ||
| Yes | 74 | 1.71±2.71 | ||
| No | 45 | 2.34±4.80 | ||
| Tumor location | F=0.565a | 0.570 | ||
| Upper | 14 | 1.09±0.83 | ||
| Middle | 69 | 1.93±3.99 | ||
| Lower | 36 | 2.31±3.59 | ||
| Tumor grade | F=1.481a | 0.232 | ||
| Well | 23 | 2.44±2.99 | ||
| Moderately | 64 | 1.42±2.28 | ||
| Poorly | 32 | 2.64±5.67 | ||
| T status | F=4.048a | 0.009* | ||
| T1 | 8 | 0.72±0.59 | ||
| T2 | 20 | 1.12±1.55 | ||
| T3 | 62 | 1.46±1.64 | ||
| T4 | 29 | 3.88±6.55 | ||
| N status | −0.789 | 0.431 | ||
| N0 | 54 | 1.66±4.26 | ||
| N1–3 | 65 | 2.19±3.03 | ||
| TNM stage | −2.974 | 0.004* | ||
| I–II | 53 | 0.97±1.02 | ||
| III | 66 | 2.73±4.67 | ||
a, one-way ANOVA test was utilized; *, comparison of miR-145-5p expression between or among each variable is statistical significance. ESCC, esophageal squamous cell carcinoma; SD, standard deviation; t, Student’s t-test; ANOVA, analysis of variance.
Patients and patient-follow-up
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The patients with ESCC collected from public databases all received treatment based on the guideline rules. The clinical characteristics of patients, including gender, age, drinking history, tumor location, tumor grade and TNM stage, were included in the analysis. The primary outcome of the present study was overall survival (OS), defined as the time from curative resection to the time of death. Relatively, the secondary outcome was DFS, calculated from the date of radical operation to the date of disease recurrence or distant metastases diagnosed.
Cell culture and virus infection
Eca109 was cultured with 1640 medium (Shanghai Yuanpei Biotechnology Co., Ltd., China), in which 10% fetal bovine serum (GIBCO) was added and placed in an incubator containing 5% CO2 at 37 °C.
When the cell growth reached the fusion degree of 60–80%, Eca109 cells were infected with miR-145-5p control lentivirus and miR-145-5p overexpression lentivirus. Polybrane was added in the ratio of 1:1,000 throughout the infection to enhance the fusion ability of virus particles and cell membrane.
RNA extraction and real-time polymerase chain reaction (RT-PCR)
Trizol Kit (Life Technologies) was used to extract total RNA from cell lines. cDNA was synthesized by reverse transcription PCR using prime scripttm RT reagent kit (Takra). The reaction conditions are: 37 °C for 30 min, 85 °C for 15 s, 4 °C. After reverse transcription, TB green premix ex taqtm II (Takra) was used for fluorescence quantitative PCR for 15 minutes. The reaction conditions were 95 °C for 15 seconds, 56 °C for 15 seconds, 68 °C for 30 seconds, and 40 cycles. Relative miR expression levels were calculated using the 2−ΔΔCt.
CCK-8 and Transwell assays
CCK-8 Kit (cell counting kit-8, biosharp, China) was used to detect cell proliferation. Cell suspension with the density of 103 cells/well was inoculated in 96 well plate and placed in 5% CO2 cell incubator at 37 °C. On the 1st, 2nd, 3rd and 4th day after the cells adhered to the wall, 10 µL CCK-8 reagent was added to 100 µL medium in each well respectively incubate for 1 h. Then the absorbance value was measured at the wavelength of 450 nm. Repeat the experiment three times.
The invasion ability of cells in vitro was detected by Transwell chamber invasion test. Cells were starved in serum-free RPMI-1640 medium for 24 hours. Adjust the number of logarithmic cells to 1–1.5 million cells/mL, and add 0.2-mL cell suspension to 24 well cross well chamber. The lower chamber was filled with 0.5 mL of RPMI-1640 medium containing fetal bovine serum. For the invasion experiment, the upper chamber with an 8-µm pore size was coated with Matrigel according to the manufacturer’s instructions.
Cells were fixed with paraformaldehyde and stained with 0.1% crystal violet. Remove the cells on the upper surface of the membrane with a cotton swab. Finally, under the microscope (×200) count the cells and calculate the average using five randomly selected fields.
miR-target interactions (MTIs)
The miRTarBase (http://mirtarbase.cuhk.edu.cn) is a comprehensive collection of MTIs, which are collected by manually surveying the pertinent literature after the natural language processing of the text systematically to filter research articles related to the functional studies of miRs. Generally, the collected MTIs are validated experimentally. Therefore, we used the miRTarBase to identify targets of miR-145-5p.
Functional analysis
Functional enrichment was achieved with MSigDB and the Gene Set Enrichment Analysis (GSEA) method. GSEA4.1.0 software was used for our analysis. The mRNA and corresponding miR-145-5p expression data of 81 ESCC patients were imported into the GSEA software, and the parameters were set as follows: Gene sets database: c2.cp.kegg.v7.2.symbols.gmt, Number of permutations: 1,000, Phenotype labels: miR-145-5p, Collapse dataset to gene symbols: No collapse, Enrichment statistic: weighted, Metric for ranking genes: Pearson. The REACTOME knowledgebase (https://reactome.org/) provides an integrated view of the molecular details of human biological processes that range from metabolism to DNA replication and repair to signaling cascades and is an extended version of a classic metabolic map in a single consistent data model.
Protein-protein interaction analyses
STRING (https://string-db.org/) is a database of known and predicted protein‒protein interactions that stem from computational predictions, from knowledge transfers between organisms, and from interactions aggregated from other (primary) databases. We applied the STRING database to analyze the interaction network of targets of miR-145-5p. Hub genes were obtained using Cytoscape (combined score >0.6, degree >10).
TIMER database analysis
TIMER2.0 (http://timer.cistrome.org/), which provides immune infiltrates’ abundances estimated by multiple immune deconvolution methods (including TIMER, CIBERSORT, QUANTISEQ, XCELL, MCPCOUNTER, and EPIC algorithms) is a comprehensive resource for the systematic analysis of immune infiltrates across diverse cancer types. The mRNA data of 81 ESCC patients were imported to calculate the immune infiltrates’ abundances for investigation of the role of miR-145-5p in the immune microenvironment.
Statistical analysis
All statistical analyses were performed using SPSS Statistics 20.0 software(IBM Corp., Armonk, NY, USA), GraphPad Prism (version 8.0.1.244), and R software (version 3.6.3). Classified variables are shown as frequencies. The best cut-off value was determined by X-Tile software. The relationship between the miR-145-5p expression level and survival rates was obtained with the Kaplan-Meier survival approach, along with the log-rank (Mantel-Cox) test. Pearson correlation coefficients were used to analyze the correlations between miR-145-5p expression and other gene or immune infiltration levels in ESCC. All statistical tests with a P value of less than 0.05 were considered significant.
Results
Significant downregulation of the expression level of miR-145-5p in ESCC tissues
We downloaded the miR data and the clinical materials of ESCC patients from the TCGA, GSE43732, and GSE13937 databases. The analysis showed that the miR-145-5p level was lower in ESCC tissues than in adjacent noncancerous tissues in the TCGA database (P<0.05) and GSE43732 database (P<0.0001), as well as the GSE13937 database (P<0.0001) (Figure 1A-1C).

Relationships between the miR-145-5p levels in ESCC tissues and clinical parameters
We studied the relationship between the miR-145-5p levels and the clinical parameters in the TCGA and GSE43732 databases (Tables 1,2). The miR-145-5p level was significantly associated with the M stages in the TCGA database. In the cancer tissues of ESCC patients with stage M1 disease, the miR-145-5p levels were significantly higher in TCGA. Although the miR-145-5p level was significantly associated with the T stages and TNM stages in GSE43732 (P<0.05), this correlation was not significant in TCGA. In the cancer tissues of ESCC patients with a higher T stage or TNM stage, the miR-145-5p levels were significantly higher in GSE43732. However, no other clinical parameters were associated with the miR-145-5p levels with statistical significance, including the demographics of gender, age, or alcohol or the tumor location, tumor grade, or presence of lymph node metastasis (P>0.05).
The survival rate of patients with higher miR-145-5p levels was significantly lower than that of patients with lower levels of miR-145-5p
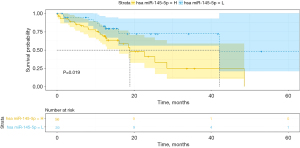
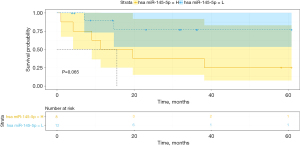
The Kaplan-Meier analysis results are shown in Figure 2A-2C. The patients with miR-145-5p levels lower than the best cut-off value were assigned to the low-expression group; the other patients were assigned to the high-expression group. An increased miR-145-5p expression was significantly associated with decreased OS and this finding remained significant in the multivariate analysis (HR, 0.206; 95% CI, 0.055–0.766, P=0.018) in TCGA (Table 3). The OS rate was significantly higher in the miR-145-5p low-expression group in TCGA (P<0.05, cut-off value =10.44), GSE43732 (P<0.01, cut-off value =1.00), as well as GSE13937 (P<0.05, cut-off value =13.79). What is more, the disease-free survival rate was also significantly higher in the miR-145-5p low-expression group in TCGA (P<0.05, cut-off value =10.86) (Figure 3). Among 20 patients receiving neoadjuvant chemoradiotherapy in GSE13937, patients with high miR-145-5p expression had a higher OS rate, although the difference was not significant due to the lack of enough samples (P=0.065, cut-off value =13.64) (Figure 4).
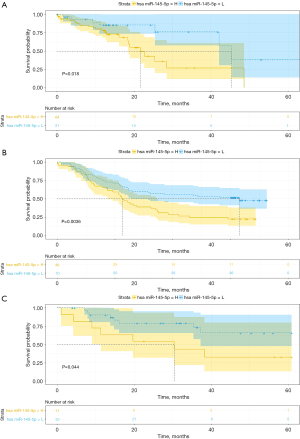
Table 3
| Parameter | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | ||
| Gender | 1.281 (0.511–3.212) | 0.597 | |||
| Male | |||||
| Female | |||||
| Age | 1.116 (0.539–2.309) | 0.767 | |||
| ≥60 | |||||
| <60 | |||||
| Alcohol | 0.579 (0.257–1.304) | 0.187 | |||
| Yes | |||||
| No | |||||
| Location | 0.090 | ||||
| Proximal | |||||
| Mid | 0.466 (0.059–3.661) | 0.468 | |||
| Distal | 2.195 (0.995–4.843) | 0.052 | |||
| G status | 0.022* | 0.023 | |||
| G1 | |||||
| G2 | 0.345 (0.107–1.111) | 0.075 | 0.265 (0.054–1.303) | 0.102 | |
| G3 | 0.334 (0.149–0.751) | 0.008* | 0.270 (0.101–0.721) | 0.009* | |
| M status | 0.444 (0.132–1.492) | 0.189 | |||
| M0 | |||||
| M1 | |||||
| T status | 1.251 (0.607–2.578) | 0.543 | |||
| T1–2 | |||||
| T3–4 | |||||
| N status | 0.707 (0.340–1.468) | 0.352 | 0.276 (0.106–0.721) | 0.009* | |
| N0 | |||||
| N1–2 | |||||
| Stage | 0.729 (0.331–1.607) | 0.434 | |||
| I–II | |||||
| III–IV | |||||
| miR-145-5p expression status | 0.359 (0.149–0.863) | 0.022* | 0.206 (0.055–0.766) | 0.018* | |
| <10.44 | |||||
| ≥10.44 | |||||
*, P<0.05. TCGA, The Cancer Genome Atlas; HR, hazard ratio; CI, confidence interval.
Upregulated expression of miR-145-5p inhibited the proliferation, migration and invasion of Eca109 cells
The down-regulation of mir-145-5p level in ESCC indicates that the miR plays a role of tumor inhibitor in ESCC. We infected Eca109 cells with miR-145-5p overexpression lentivirus and evaluated the effect on cell biological function in vitro. TaqMan RT-PCR showed that the expression of miR-145-5p was significantly up-regulated in Eca109 cell (Figure 5A). To identify whether miR-30b-5p has an effect on the progress of ESCC, we performed CCK8 and Transwell experiments. Up regulation of miR-145-5p attenuated the proliferation, migration and invasion of Eca109 cells (Figure 5B-5D). These results suggest that miR-145-5p acts as an inhibitor in ESCC.

Several tumor and immune-related pathways may be involved in the upregulation of miR-145-5p
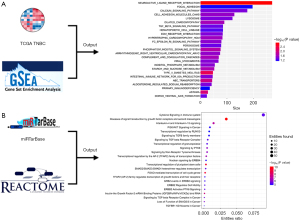
Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was performed to comprehensively study the interaction between miR-145-5p and its function. Functional enrichment analysis revealed that the miR-145-5p was enriched in inflammation (including the Fc epsilon RI signaling pathway, complement and coagulation cascades, asthma pathways, and others), metastasis (including focal adhesions, cell adhesion molecules, extracellular matrix receptor interaction pathways, and others), and metabolism (including starch and sucrose metabolism pathways and others). The top 25 pathways are shown in Figure 6A. We used another approach to further confirm which biofunctions were correlated with miR-145-5p. REACTOME, which was a functional enrichment tool, was used to align the targets from the miRTarBase and the biofunctions of miR-145-5p, and the results showed that miR-145-5p was correlated with the cancer, immune system, metabolism, and signal transduction. The top 25 functional pathways were ranked with the entities ratio (Figure 6B). These results suggested that miR-145-5p is most involved in inflammation and cancer metastasis. What is more, we noticed that the transforming growth factor-beta (TGF-β) signaling pathway, which had been reported to play an important role in the immune microenvironment and tumor metastasis in previous studies (13,14), was significantly enriched in both methods.
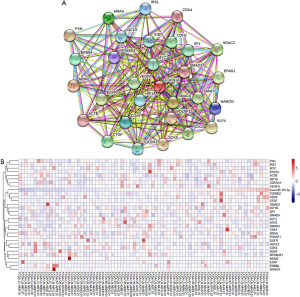
The levels of TGFBR2, TCGF, and CDH2 were significantly correlated with miR-145-5p
To obtain the key targets, the Search Tool for the Retrieval of Interacting Genes (STRING) was applied to analyze the interaction network of targets from the miRTarBase. Thirty genes were defined as hub genes (Figure 7A) using Cytoscape; the expression levels in TCGA are shown in Figure 7B. Pearson correlation analysis was performed to clarify the relationship between miR-145-5p and the hub genes in ESCC, and the result showed that the TGFBR2 levels (P=0.0033, r=0.3225), TCGF levels (P=0.0001, r=0.4137), and CDH2 levels (P<0.0001, r=0.4305) were most correlated with miR-145-5p (Figure 8A-8C).
Macrophage infiltration level correlates with miR-145-5p in ESCC
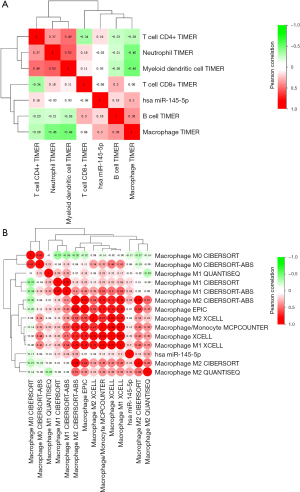
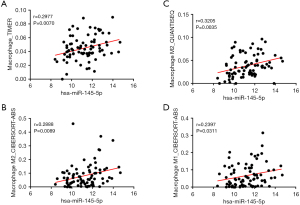
Previous studies have indicated that miR-145-5p could be an effective biomarker in predicting the response to immunochemotherapy or the enhancements (7,8). In order to investigate the crucial role of miR-145-5p in tumor immunity, we analyzed the relationship between miR-145-5p expression and the immune infiltration level in ESCC. The result of the Pearson correlation analysis is shown in Figure 9A. To confirm the relation between macrophage infiltration level and miR-145-5p, we used multiple algorithms for further analysis. The result of the Pearson correlation analysis is shown in Figure 9B. We noticed that the level of macrophage infiltration was most significantly correlated with miR-145-5p (r=0.2977, P=0.0070) (Figure 10A). However, the B-cell, CD4+ T-cell, CD8+ T-cell, neutrophil, and myeloid dendritic cell infiltration levels showed no significant connection with miR-145-5p expression in ESCC (P>0.05). In addition, the miR-145-5p has a positive relationship to the M2 macrophage infiltration level in CIBERSORT-ABS (r=0.2888, P=0.0089) and QUANTISEQ (r=0.3205, P=0.0035) (Figure 10B,10C) and is correlated with an increased M1 macrophage infiltration level in CIBERSORT-ABS (r=0.2397, P=0.0311) (Figure 10D). Only the M0 macrophage infiltration level shows no significant connection with miR-145-5p expression (P>0.05). Our work demonstrated the important role of miR-145-5p in the macrophage infiltration level, particularly for M2 macrophages, in ESCC.
Correlation between miR-145-5p and gene markers of macrophages
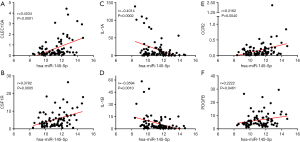
To validate the above findings, we consulted previous research and selected the gene markers of different functional macrophages, including M1, M2, and tumor-associated macrophages, for further analysis (15). The miR-145-5p had positive relationships to Clec10A/CD301 (r=0.4534, P<0.0001) and CSF1R/CD115 (r=0.3782, P=0.0005) (Figure 11A,11B), which had been reported as key markers of M2 macrophages in a previous study. Surprisingly, miR-145-5p was negatively correlated with key markers of M1 macrophages, such as IL-1A (r=‒0.4014, P=0.0002) and IL-1B (r=‒0.3594, P=0.0010) (Figure 11C,11D). These findings suggested that the upregulation of miR-145-5p promotes macrophage polarization to M2. One article reported the underlying relationship between miR-145-5p and M2 macrophages (16), and our work first verified their close link in ESCC by immune infiltration analysis. We noticed that miR-145-5p has significant interactions with the key genes of tumor-associated macrophages, such as CCR2 (r=0.3162, P=0.0040) and PDGFB (r=0.2222, P=0.0461) (Figure 11E,11F); this shows its potential value and the worth of further investigating its underlying mechanism.
Discussion
ESCC is a common malignant carcinoma with a poor prognosis, and its treatment requires surgery, chemotherapy, and/or radiotherapy, all of which have severe side effects. Since genetic diseases and immune disorders in tumor are regarded as promoting factors during tumorigenesis and development including ESCC, targeting tumor microenvironment and tumor cells seems to be a significant direction for research on which treatments could improve the prognosis of patients with ESCC (17,18). In the current study, we investigated the expression of miR-145-5p in ESCC and elucidated its relationship with clinical parameters and immunochemotherapy.
According to the results of our study, the expression level of miR-145-5p was significantly lower in cancer tissues compared with adjacent noncancerous tissues in patients with ESCC, and this may suggest an association of miR-145-5p with the occurrence of ESCC. Additionally, functional experiments showed that miR-145-5p attenuated proliferation, migration and invasion in the Eca109 cell line. In fact, much evidence has revealed that miR-145-5p functioned as a tumor suppressor and that the expression levels of miR-145-5p were extremely downregulated in certain types of cancers to avoid antioncogenic functions (10,17,19-21). The results of our study also show that the miR-145-5p level is significantly associated with the progression of ESCC. In analyzing patients’ data, the expression level of miR-145-5p was upregulated in patients with an advanced stage of ESCC. The associations between miR-145-5p and the M, T, and TNM stages were supported in our analysis of the TCGA and GEO43732 data. A previous study reported that a high level of miR-145-5p was positively correlated with the recurrence of metastasis, and this was consistent with our conclusions (11).
In addition, high expression levels of miR-145-5p predict a poor prognosis for patients with ESCC. The results are consistent for both the TCGA and GEO databases. The close relationship between miR-145-5p and the prognosis for ESCC may suggest the potential application of miR-145-5p as a predictor of prognosis among patients with ESCC. Previous studies showed that a high level of miR-145-5p predicts a poor response to neoadjuvant chemotherapy (7,8), and our study first suggested that a high level of miR-145-5p was associated with a poor prognosis for patients undergoing neoadjuvant chemotherapy. However, the statistical difference was not significant in our study due to the lack of enough samples. Thus, further studies with a large cohort of patients are needed in the future.
We further performed functional enrichment analysis and immune infiltration analysis to investigate the relationship between miR-145-5p and the immune infiltration level in ESCC. The analysis of the signal pathways associated with miR-145-5p indicated that the inflammation and cancer-related pathways of inflammation, metastasis, and metabolism were most significantly enriched in ESCC. In addition, we noticed a close relationship between miR-145-5p and the TGF-β signaling pathway. As previously reported, TGF-β can be secreted by tumor-associated macrophages and cancer-associated fibroblasts, as well as cancer cells, and is constitutively active in many tumors (22,23). Mounting numbers of studies have established that the TGF-β pathway plays a vital role in regulating the production of the extracellular matrix and tumor metastasis and is an essential factor in cancer development (24-26). The duration and intensity of the TGF-β signaling pathway activity are critical determinants of the biological responses of TGF-β family members. Several genes of the TGF-β1 pathway were found to have a possible predictive value for ESCC in patients receiving neoadjuvant chemoradiotherapy (27). Our current research suggested that miR-145-5p may be a key regulator of the TGF-β signaling pathway in ESCC. Moreover, the miR-145-5p level was positively correlated with the TGF-β signaling pathway‒associated genes TGFBR2, CTGF, and CDH2. CTGF was shown to act as a downstream factor of the TGF-β pathway, and it enhanced the tumor cellular invasiveness and diffusion in ESCC (28). Similarly, TGF-β‒activated CDH2 could promote epithelial-mesenchymal transition and cancer cell invasion and migration (29).
In addition, immune infiltration analysis showed that miR-145-5p was positively correlated with the M macrophage infiltration level and the gene markers of M2 macrophages and tumor-associated macrophages in ESCC. Several studies reported that the upregulation of miR-145-5p promotes the polarization of M2 macrophages (8,16,30), which was correlated with high microvessel density and matrix breakdown, and that this may contribute to angiogenesis and tumor aggressiveness, promoting the progression of ESCC (31). To the best of our knowledge, this is the first time the association between miR-145-5p and macrophage infiltration has been reported in ESCC. ESCC was enriched in immunosuppressive cell populations, including Tregs, exhausted CD8 T, CD4 T and NK cells, M2 macrophages, and tDCs. All these immune-inhibitory cells may contribute to immune escape and promote tumor progression. Recent studies have shown that the interaction between macrophages and Treg through ligand receptor interaction may contribute to immunosuppressive status and disease progression (32). Macrophages are the main components of the immune infiltrate of the tumor microenvironment and can produce a variety of phenotypes in different microenvironments, including classically activated (M1) and alternately activated (M2) macrophages (33). M1-like cells are involved in pathogen clearance and proinflammatory responses, and M2-like cells promote tissue remodeling and anti-inflammatory processes and are related to tumor progression (15). Macrophages that infiltrate tumor tissues, called tumor-associated macrophages, are associated with tumor progression and metastasis. Infiltration of tumor-associated macrophages, especially M2 macrophages, was reported to be associated with a poor response to chemotherapy and a poor prognosis for patients with esophageal cancer (34). Our study revealed that miR-145-5p plays a crucial role in tumor-associated macrophages.
Interestingly, our findings suggested that miR-145-5p has a dual function. It plays a tumor-suppressive role by inhibiting cancer cell proliferation, migration and invasion, and epithelial-mesenchymal transition in multiple cancer types, including ESCC, and it is also positively correlated with high tumor stages and a poor prognosis in ESCC. In fact, these two special and seemingly opposite trends are not contradictory. Recent studies have provided some insights that may explain the underlying mechanisms (8,35). On the one hand, a low expression of miR-145-5p in esophageal cancer cells can reduce its antitumor role. On the other hand, a high level of miR-145-5p promotes tumor-associated macrophage infiltration and macrophage polarization to M2 via ESCC-derived extracellular vesicles and leads to tumor progression and a poor prognosis while reducing the response to chemoradiotherapy via several signaling pathways, including the TGF-β signaling pathway.
Conclusions
In summary, our research demonstrated that miR-145-5p acts as a tumor suppressor microRNA and plays an important role in the high M2-like tumor-associated macrophage infiltration of ESCC. High miR-145-5p levels in ESCC samples are associated with advanced stages and a poor prognosis, and they predict a poor response to chemoradiotherapy. The miR-145-5p may be a potential target for immunotherapy and a predictor of the prognosis for the treatment of immunotherapy. However, more studies are needed for further investigation.
Acknowledgments
Funding: This study was funded by the Zhejiang Provincial Department of Medical and Health Science and Technology (grant number 2015KYA239), Taizhou science and technology planning project (grant number 1802ky01) and Shaoxing People’s Hospital Youth Scientific Research Fund Project (grant number 2020YB27).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-294/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-294/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wu X, Zhang H, Sui Z, et al. The biological role of the CXCL12/CXCR4 axis in esophageal squamous cell carcinoma. Cancer Biol Med 2021;18:401-10. [Crossref] [PubMed]
- Veen L, Hulshof M, Jimenez CR, et al. TGF-β and PD-L1 inhibition combined with definitive chemoradiotherapy in esophageal squamous cell carcinoma: A phase II clinical trial (NCT04595149). J Clin Oncol 2021;39:TPS4154. [Crossref]
- Cui M, Yao X, Lin Y, et al. Interactive functions of microRNAs in the miR-23a-27a-24-2 cluster and the potential for targeted therapy in cancer. J Cell Physiol 2020;235:6-16. [Crossref] [PubMed]
- Borchardt H, Ewe A, Morawski M, et al. miR24-3p activity after delivery into pancreatic carcinoma cell lines exerts profound tumor-inhibitory effects through distinct pathways of apoptosis and autophagy induction. Cancer Lett 2021;503:174-84. [Crossref] [PubMed]
- Skogstrøm KB, Sletten M, Tinholt M, et al. PO-70 Anti-tumour miR-7-5p as a novel regulator of TFPI in liver cancer. Thrombosis Research 2021;200:S56. [Crossref]
- Cui Z, Sun Q, Yan W, et al. The role of miR-320a and its target gene GMEB1 in epithelial-mesenchymal transition and invasion of colorectal cancer. J Gene Med 2021; Epub ahead of print. [Crossref] [PubMed]
- Wen J, Luo K, Liu H, et al. MiRNA Expression Analysis of Pretreatment Biopsies Predicts the Pathological Response of Esophageal Squamous Cell Carcinomas to Neoadjuvant Chemoradiotherapy. Ann Surg 2016;263:942-8. [Crossref] [PubMed]
- García-García F, Salinas-Vera YM, García-Vázquez R, et al. miR-145-5p is associated with pathological complete response to neoadjuvant chemotherapy and impairs cell proliferation by targeting TGFβR2 in breast cancer. Oncol Rep 2019;41:3527-34. [PubMed]
- Zheng TL, Li DP, He ZF, et al. miR-145 sensitizes esophageal squamous cell carcinoma to cisplatin through directly inhibiting PI3K/AKT signaling pathway. Cancer Cell Int 2019;19:250. [Crossref] [PubMed]
- Fan S, Chen P, Li S. miR-145-5p Inhibits the Proliferation, Migration, and Invasion of Esophageal Carcinoma Cells by Targeting ABRACL. Biomed Res Int 2021;2021:6692544. [Crossref] [PubMed]
- Tabrizi M, Khalili M, Vasei M, et al. Evaluating the miR-302b and miR-145 expression in formalin-fixed paraffin-embedded samples of esophageal squamous cell carcinoma. Arch Iran Med 2015;18:173-8. [PubMed]
- Hayward SW. Immunotherapeutic Response in Tumors Is Affected by Microenvironmental ROS. Cancer Res 2020;80:1799-800. [Crossref] [PubMed]
- Huang X, Zhang G, Liang T. Cancer environmental immunotherapy: starving tumor cell to death by targeting TGFB on immune cell. J Immunother Cancer 2021;9:e002823. [Crossref] [PubMed]
- Hussain SM, Kansal RG, Alvarez MA, et al. Role of TGF-β in pancreatic ductal adenocarcinoma progression and PD-L1 expression. Cell Oncol (Dordr) 2021;44:673-87. [Crossref] [PubMed]
- Li J, Xie Y, Wang X, et al. Prognostic impact of tumor-associated macrophage infiltration in esophageal cancer: a meta-analysis. Future Oncol 2019;15:2303-17. [Crossref] [PubMed]
- Su D. Up-regulation of MiR-145-5p promotes the growth and migration in LPS-treated HUVECs through inducing macrophage polarization to M2. J Recept Signal Transduct Res 2021;41:434-41. [Crossref] [PubMed]
- Ding Y, Zhang C, Zhang J, et al. miR-145 inhibits proliferation and migration of breast cancer cells by directly or indirectly regulating TGF-β1 expression. Int J Oncol 2017;50:1701-10. [Crossref] [PubMed]
- Guo JC, Hsu CL, Huang YL, et al. Association of B cells in tumor microenvironment (TME) with clinical benefit to programmed cell death protein-1 (PD-1) blockade therapy in esophageal squamous cell carcinoma (ESCC). J Clin Oncol 2021;39:abstr 237.
- Li H, Zhao S, Chen X, et al. MiR-145 modulates the radiosensitivity of non-small cell lung cancer cells by suppression of TMOD3. Carcinogenesis 2022;43:288-96. [Crossref] [PubMed]
- Hang W, Feng Y, Sang Z, et al. Downregulation of miR-145-5p in cancer cells and their derived exosomes may contribute to the development of ovarian cancer by targeting CT. Int J Mol Med 2019;43:256-66. [PubMed]
- Bahreini F, Saidijam M, Mousivand Z, et al. Assessment of lncRNA DANCR, miR-145-5p and NRAS axis as biomarkers for the diagnosis of colorectal cancer. Mol Biol Rep 2021;48:3541-7. [Crossref] [PubMed]
- Ning J, Ye Y, Bu D, et al. Imbalance of TGF-β1/BMP-7 pathways induced by M2-polarized macrophages promotes hepatocellular carcinoma aggressiveness. Mol Ther 2021;29:2067-87. [Crossref] [PubMed]
- Liu C, Zhang W, Wang J, et al. Tumor-associated macrophage-derived transforming growth factor-β promotes colorectal cancer progression through HIF1-TRIB3 signaling. Cancer Sci 2021;112:4198-207. [Crossref] [PubMed]
- Benjamin DJ, Lyou Y. Advances in Immunotherapy and the TGF-β Resistance Pathway in Metastatic Bladder Cancer. Cancers (Basel) 2021;13:5724. [Crossref] [PubMed]
- Hong Y, Li X, Zhu J. LSD1-mediated stabilization of SEPT6 protein activates the TGF-β1 pathway and regulates non-small-cell lung cancer metastasis. Cancer Gene Ther 2022;29:189-201. [Crossref] [PubMed]
- Yang Y, Ye WL, Zhang RN, et al. The Role of TGF-β Signaling Pathways in Cancer and Its Potential as a Therapeutic Target. Evid Based Complement Alternat Med 2021;2021:6675208. [Crossref] [PubMed]
- Smith JJ, Wasserman I, Milgrom SA, et al. Single Nucleotide Polymorphism TGFβ1 R25P Correlates with Acute Toxicity during Neoadjuvant Chemoradiotherapy in Rectal Cancer Patients. Int J Radiat Oncol Biol Phys 2017;97:924-30. [Crossref] [PubMed]
- Xie JJ, Xu LY, Wu JY, et al. Involvement of CYR61 and CTGF in the fascin-mediated proliferation and invasiveness of esophageal squamous cell carcinomas cells. Am J Pathol 2010;176:939-51. [Crossref] [PubMed]
- Wang L, Peng Q, Sai B, et al. Ligand-independent EphB1 signaling mediates TGF-β-activated CDH2 and promotes lung cancer cell invasion and migration. J Cancer 2020;11:4123-31. [Crossref] [PubMed]
- Shinohara H, Kuranaga Y, Kumazaki M, et al. Regulated Polarization of Tumor-Associated Macrophages by miR-145 via Colorectal Cancer-Derived Extracellular Vesicles. J Immunol 2017;199:1505-15. [Crossref] [PubMed]
- Nakajima S, Mimura K, Saito K, et al. Neoadjuvant Chemotherapy Induces IL34 Signaling and Promotes Chemoresistance via Tumor-Associated Macrophage Polarization in Esophageal Squamous Cell Carcinoma. Mol Cancer Res 2021;19:1085-95. [Crossref] [PubMed]
- Zheng Y, Chen Z, Han Y, et al. Immune suppressive landscape in the human esophageal squamous cell carcinoma microenvironment. Nat Commun 2020;11:6268. [Crossref] [PubMed]
- Eftimie R, Barelle C. Mathematical investigation of innate immune responses to lung cancer: The role of macrophages with mixed phenotypes. J Theor Biol 2021;524:110739. [Crossref] [PubMed]
- Tzao C, Cheng LY, Chang CC. 492 tumor associated macrophage promotes epithelial-to-mesenchymal transition in esophageal squamous cell cancer. Diseases of the Esophagus 2021. Available online:
10.1093/dote/doab052.492 10.1093/dote/doab052.492 - Cao Y, Jiao N, Sun T, et al. CXCL11 Correlates With Antitumor Immunity and an Improved Prognosis in Colon Cancer. Front Cell Dev Biol 2021;9:646252. [Crossref] [PubMed]


