A systematic review and meta-analysis of circulating cell-free DNA as a diagnostic biomarker for non-small cell lung cancer
Introduction
Lung cancer has one of the highest incidence rates among all malignant tumors, with 1.6 million morbidities and 1.38 million deaths annually (1). Non-small cell lung cancer (NSCLC), with an alarming incidence, accounting for 85% of lung cancers (2), with the majority of patients already at an advanced stage at diagnosis, and usually less than 5% of patients survive 5 years (3-5). However, if NSCLC is diagnosed early, the survival rate of patients after surgical removal of the tumor can exceed 80% (6,7). At present, the recommended low-dose computed tomography (LDCT) screening method for early detection of lung cancer still encounters some challenges (8,9), and there is a lack of clinically good biomarkers for early diagnosis of NSCLC.
With the development of precision cancer medicine, genome analysis has attracted increasing attention for the diagnosis and treatment of tumors. Genome sequencing of tumor biopsy specimens is currently a relatively advanced auxiliary diagnostic technology. It has proven its application value in confirming treatment efficacy and predicting treatment response (10). At the same time, it can also provide reference value in terms of disease activity and drug resistance (11,12). For example, the sequencing of breast cancer is of great reference in the selection of treatment options, especially aromatase inhibitors (10). However, genotyping requires biopsy specimens from cancer patients, and its availability is limited (13,14). At the same time, biopsy may not be able to obtain enough tumor tissue in some patients, and there may also be operational risks and false negative results.
Circulating cell-free DNA (cfDNA) in plasma, possibly originate from necrosis, apoptosis, and/or macrophage digestion. Circulating cfDNA has been regarded as a new biomarker for the diagnosis, treatment and prognosis of malignant tumors (15). Even a very small tumor will release enough cfDNA in the blood, which is well below the detection limit of radiological methods (16). Most of the circulating cfDNA is excreted from the cell after being cut by endonuclease (17). In healthy individuals, circulating cfDNA enters the blood in two ways, through the circulation of lymphocytes and through apoptosis of other nucleated cells (18). In cancer patients, circulating cfDNA is produced after tumor necrosis, and includes the lysis of circulating malignant cells or micro metastasis (19).
The value of cfDNA in early-stage NSCLC is more reflected, such as tumor discovery, tumor burden inspection, and minimal residual tumor monitoring. CfDNA can be obtained through patient serum or minimally invasive surgery, and reflects genetic changes in tumor tissues, and thus, cfDNA testing is considered an important method for the diagnosis of NSCLC. However, there is still controversy about its accuracy in diagnosing NSCLC (18-21). For example, a prospective study reported that the baseline cfDNA levels may be not associated with progression-free survival (PFS) or overall survival (OS), neither were changes in cfDNA (15). While other scholars showed a significant relationship between low DNA integrity and OS between those patients with longer survival for patients with low levels (22). The purpose of this work is to further support that circulating cfDNA can be used as a biomarker for the diagnosis of NSCLC. We present the following article in accordance with the PRISMA-DTA reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-646/rc).
Methods
Search strategy
We performed a literature search of China National Knowledge Infrastructure (CNKI), Wanfang, and VIP and English (PubMed, Cochrane Central Register of Controlled Trials, Embase, and Web of Science) biomedical databases using the following search terms: lung tumors, lung cancer, NSCLC, biomarkers, circulating cfDNA, cfDNA, ctDNA, circulating cell-free tumor DNA, diagnosis, prognosis, and monitoring. The retrieval period was set from the opening of the database to September 2021.
Inclusion and exclusion criteria
According to the PICOS (patients, intervention, comparison, outcomes, and study design) principles, the inclusion criteria were as follows: (I) NSCLC patients must be diagnosed by histopathology or cytology; (II) diagnosis intervention by circulating cfDNA was the same as histopathology or cytology; (III) patients aged ≥18 years in the involved studies; (IV) studies that reported on at least 10 lung cancers in the study population; (V) articles using circulating cfDNA as a biomarker to diagnosis of NSCLC patients, the participants should include both the patients and non-patient subjects; (VI) article outcome data could be completely extracted.
The exclusion criteria were as follows: (I) tumor tissue and blood samples are not matched; (II) the population of the research report is the same; and (III) no circulating cfDNA was used to diagnose NSCLC.
Literature screening and data extraction
Two review authors were selected to read, analyze the abstracts of the studies obtained according to the inclusion criteria. In the case of different opinions among reviewers, a third reviewer is recommended to review the manuscript independently and blindly. After completing the review, review authors independently reproduced useful data from the screened articles, including first author, country of study, year of publication, sample size, and true-positive, true-negative, false-positive, and false-negative forms. There are different opinions during data replication, and a consensus is reached through discussion. Articles for which no valid data could be obtained were excluded.
Risk of bias assessment
The research quality assessment scale QUADAS (Quality Assessment of Diagnostic Accuracy Studies)-2 recommended by the Cochrane Collaborative Organization was used to evaluate the methodological quality of the included research. Patient selection for the study, reporting index tests, reference content criteria, flow and timing were all included in the assessment. Risk of bias analysis was performed for each index, and the first three indexes were evaluated according to the question of suitability. The methodological quality indicator was rated as “low risk”, “high risk”, or “unclear”. Publication bias was assessed visually by funnel plots in RevMan.
Statistical analysis
The χ2 test was used to test statistical heterogeneity, and the I2 statistic was used to evaluate the degree of variation that could be attributed to the statistical heterogeneity between the trials. An I2<50% indicates low heterogeneity, while an I2>50% represents significant heterogeneity. At first, a fixed-effects model was used to drive the overall effect sizes. If there was significant between-studies heterogeneity, the random-effects model (DerSimonian-Laird) was applied as an alternative. In this study, the random effects model was used when I2>30%; otherwise, the fixed effects model was applied. The forest plot was used to display the sensitivity and specificity of the indicators in this study, and the 95% confidence interval (CI) was calculated. For the accuracy of each result, a summary receiving operating characteristic (SROC) curve and the respective area under the curve (AUC) were constructed for analysis. The positive likelihood ratio, negative likelihood ratio, and diagnostic odds ratio (DOR) were also calculated and analyzed. Next, funnel plot and Egger’s test were done to assess publication bias. Statistical analysis was performed by RevMan version 5.3 software (Cochrane Collaboration, https://www.cochranelibrary.com/). P<0.05 was considered to indicate a statistically significant difference.
Results
Literature search results and study characteristics
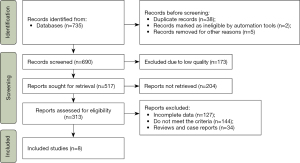
Together, 737 records were confirmed, and 47 unavailable records were eliminated. After screening, 313 articles were retrieved, including 243 English articles and 70 Chinese articles. Most articles did not meet the requirements, and eight articles were screened for further analysis. The flow chart is displayed in Figure 1. The eight articles included 618 NSCLC patients and 635 healthy subjects, and the publication period was from 2008 to 2018. The basic data of the articles, such as author, country, year, journal, research type, and number of subjects, were extracted. The basic characteristics of the eight articles are shown in Table 1.
Table 1
| Author | Country | Year | Journal | Type | Patients | Controls |
|---|---|---|---|---|---|---|
| Ulivi et al. (20) | Italy | 2008 | Thorax | Case-control | 128 | 103 |
| van der Drift et al. (21) | The Netherlands | 2010 | Lung Cancer | Prospective | 46 | 21 |
| Ulivi et al. (22) | Italy | 2013 | PLoS One | Prospective | 100 | 100 |
| Chiappetta et al. (23) | Italy | 2013 | Clin Chim Acta | Prospective | 30 | 26 |
| Catarino et al. (24) | Portugal | 2012 | PLoS One | Prospective | 104 | 205 |
| Kumar et al. (25) | India | 2010 | Lung Cancer | Prospective | 100 | 100 |
| Soliman et al. (26) | Egypt | 2018 | Biochem Biophys Rep | Prospective | 60 | 40 |
| Szpechcinski et al. (27) | Poland | 2015 | Br J Cancer | Prospective | 50 | 40 |
Risk of bias and applicability judgments
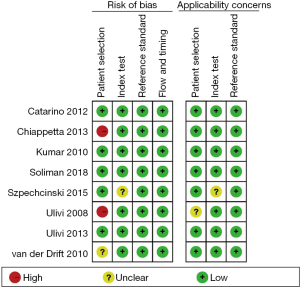
The risk of patient selection bias was unclear in one article high in two articles, and low in the remaining five articles is low. As for the index test bias, one article had an unclear risk, and the remaining seven articles were low risk. The reference standard bias and flow and timing bias of all articles were low. As for applicability bias, one article had an unclear risk of patient selection bias, one article had an unclear risk of index test bias, and the applicability bias of the remaining articles was low. The risk of bias analysis is shown in Figure 2.
Meta-analysis results
Overall analyses
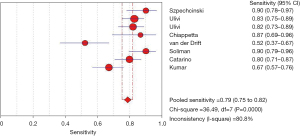
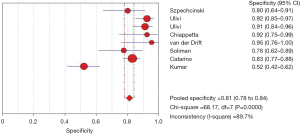
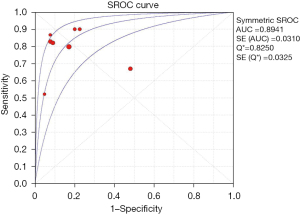
After analysis, the effect of analysis suggested circulating cfDNA as a biomarker with a pooled sensitivity of 0.79 (95% CI: 0.75–0.82) for NSCLC diagnosis and prognosis (Figure 3), and the pooled specificity was 0.81 (95% CI: 0.78–0.84) (Figure 4). In addition, the sensitivity and specificity I2 were 80.8% and 89.7%, respectively, and were thus analyzed using random effects models. The SROC showed a higher accuracy (AUC =0.8941) (Figure 5).
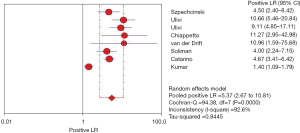
The positive likelihood ratio refers to the multiple of the correctly diagnosed disease and the wrongly diagnosed disease in the diagnostic experiment. Therefore, the greater the positive likelihood ratio, the higher the accuracy of the diagnosis. In this study, the pooled positive likelihood ratio of circulating cfDNA in the diagnosis of NSCLC was 5.37 (95% CI: 2.67–10.81), indicating that circulating cfDNA is more reliable as a diagnostic biomarker for NSCLC (Figure 6).
The negative likelihood ratio refers to the ratio of the false-negative rate and the true-negative rate detected by clinical diagnostic experiments, with smaller values indicating better diagnostic methods. The negative likelihood ratio suggests that the possibility of falsely being judged as negative is a multiple of the possibility of being correctly judged as being negative. The negative likelihood ratio summarized in this study was 0.24 (95% CI: 0.15–0.38), indicating that circulating cfDNA is more accurate as a diagnostic biomarker for NSCLC (Figure 7).

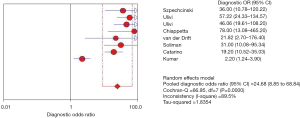
The DOR refers to the ratio of the positive likelihood ratio to the negative likelihood ratio, which is a response to the closeness of the relationship between the outcome and the disease. When this value is greater than 1, larger values denote that the diagnostic test has a better discriminatory effect; however, when this value is less than 1, healthy people are more likely to be judged as positive by the diagnostic test than the patient. Furthermore, when this value is equal to 1, this signifies that the diagnostic test unable to distinguish between healthy people and patients. The greater the DOR, the greater the ability to accurately diagnose the disease. The results of this study showed that the pooled DOR was 24.68 (95% CI: 8.85–68.84). Therefore, it can be considered that circulating cfDNA is highly authentic as a diagnostic biomarker for NSCLC (Figure 8).
Risk of bias
After quality assessment, we observed that the patient selection bias of patients was high in two articles was high (20,23), unclear in one article (21), and low in the remaining five articles (22,24-27). The risk of bias in the research index test was unclear in one article (27), and low in the remaining seven articles (20-26). The reference standard bias, and flow and time bias of all articles was low. Regarding the risk of applicability bias, one article had an unclear risk of patient selection bias (20) and index test bias (27), while the remaining articles had a low risk (Figure 9).
Discussion
NSCLC is one of the main causes of cancer-related death worldwide. Since most patients are diagnosed as advanced NSCLC, the 5-year survival rate of patients is very low (3,4,28), and the survival of NSCLC patients largely depends on early detection and diagnosis. Therefore, the development of new biomarkers that can be used in the early diagnosis of NSCLC is meaningful for patients, so that they may receive corresponding treatment as soon as possible (6,29). For patients with advanced NSCLC, only a few biopsies can be used for histological diagnosis and genetic testing, and there is a lack of tissue for genomic analysis after the initial histological diagnosis. For timely adjustment of the treatment plan when drug resistance develops, it is necessary to encourage patients to undergo a second biopsy to obtain cancer tissue for genetic analysis; however, this involves potential harm and operational risks to already weak patients. Finding new sources of cancer tissue genes may improve the diagnosis monitoring of patients.
For cancer patients, circulating serum cfDNA released by tumor cells represents a clue or evidence of the biological manifestations of cancer. Therefore, circulating serum cfDNA can be used as a suitable tool for early diagnosis monitoring. Blood-based tests are safer and easier for patients to accept (12,30). Therefore, this study aimed to analyze the published data in order to evaluate the value of circulating cfDNA for the diagnosis evaluation of NSCLC, in order to promote the use of circulating cfDNA as a reliable biomarker for the diagnosis of NSCLC patients.
We conducted a comprehensive meta-analysis to evaluate the potential significance of circulating cfDNA in the diagnosis of NSCLC patients. The overall sensitivity and specificity of circulating cfDNA as a biomarker for diagnosis of NSCLC were 0.79 (95% CI: 0.75–0.82) and 0.81 (95% CI: 0.78–0.84), respectively, and the AUC was 0.8941. The positive and negative likelihood ratios were 5.37 (95% CI: 2.67–10.81) and 0.24 (95% CI: 0.15–0.38), respectively, and the DOR was 24.68 (95% CI: 8.85–68.84). The comprehensive analysis results suggested that circulating cfDNA has high accuracy and considerable potential application value as a biomarker for the clinical diagnosis of NSCLC.
However, there are several limitations in this meta-analysis that should be noted. Firstly, some of the included studies are retrospective in design. Secondly, the sample size of included studies was relatively small. Thirdly, there was no uniform threshold standard for the included research articles. These limitations may reduce the reliability of our findings. It is necessary to implement a large-sample, multi-center, prospective cohort study to further determine the value of circulating cfDNA in the clinical diagnosis of NSCLC patients.
Conclusions
This study concluded that circulating serum cfDNA provides a promising non-invasive blood-detection prospect for the early diagnosis of NSCLC. It can be used as a reliable and accurate clinical biomarker for NSCLC and can be applied to distinguish healthy individuals from NSCLC patients. However, due to technical difficulties, the practicability and accuracy of circulating cfDNA is restricted by the testing methods to a large extent, and further development of detection technology is needed.
Acknowledgments
Funding: This work was supported by the Youth Science and Technology Project of Hebei Provincial Health Commission (No. 20190743).
Footnote
Reporting Checklist: The authors have completed the PRISMA-DTA reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-646/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-646/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Chen Z, Fillmore CM, Hammerman PS, et al. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer 2014;14:535-46. [Crossref] [PubMed]
- Spira A, Halmos B, Powell CA. Am J Respir Crit Care Med 2015;192:283-94. Update in Lung Cancer 2014. [Crossref] [PubMed]
- Tsao AS, Scagliotti GV, Bunn PA Jr, et al. Scientific Advances in Lung Cancer 2015. J Thorac Oncol 2016;11:613-38. [Crossref] [PubMed]
- Wood SL, Pernemalm M, Crosbie PA, et al. Molecular histology of lung cancer: from targets to treatments. Cancer Treat Rev 2015;41:361-75. [Crossref] [PubMed]
- Padda SK, Burt BM, Trakul N, et al. Early-stage non-small cell lung cancer: surgery, stereotactic radiosurgery, and individualized adjuvant therapy. Semin Oncol 2014;41:40-56. [Crossref] [PubMed]
- Patz EF Jr, Rossi S, Harpole DH Jr, et al. Correlation of tumor size and survival in patients with stage IA non-small cell lung cancer. Chest 2000;117:1568-71. [Crossref] [PubMed]
- Chen X, Zhou F, Li X, et al. Folate Receptor-Positive Circulating Tumor Cell Detected by LT-PCR-Based Method as a Diagnostic Biomarker for Non-Small-Cell Lung Cancer. J Thorac Oncol 2015;10:1163-71. [Crossref] [PubMed]
- Zhang R, Shao F, Wu X, et al. Value of quantitative analysis of circulating cell free DNA as a screening tool for lung cancer: a meta-analysis. Lung Cancer 2010;69:225-31. [Crossref] [PubMed]
- Ellis MJ, Ding L, Shen D, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 2012;486:353-60. [Crossref] [PubMed]
- McDermott U, Downing JR, Stratton MR. Genomics and the continuum of cancer care. N Engl J Med 2011;364:340-50. [Crossref] [PubMed]
- Murtaza M, Dawson SJ, Tsui DW, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013;497:108-12. [Crossref] [PubMed]
- De Mattos-Arruda L, Caldas C. Cell-free circulating tumour DNA as a liquid biopsy in breast cancer. Mol Oncol 2016;10:464-74. [Crossref] [PubMed]
- Mao C, Yuan JQ, Yang ZY, et al. Blood as a Substitute for Tumor Tissue in Detecting EGFR Mutations for Guiding EGFR TKIs Treatment of Nonsmall Cell Lung Cancer: A Systematic Review and Meta-Analysis. Medicine (Baltimore) 2015;94:e775. [Crossref] [PubMed]
- Li BT, Drilon A, Johnson ML, et al. A prospective study of total plasma cell-free DNA as a predictive biomarker for response to systemic therapy in patients with advanced non-small-cell lung cancers. Ann Oncol 2016;27:154-9. [Crossref] [PubMed]
- Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011;11:426-37. [Crossref] [PubMed]
- Hao TB, Shi W, Shen XJ, et al. Circulating cell-free DNA in serum as a biomarker for diagnosis and prognostic prediction of colorectal cancer. Br J Cancer 2014;111:1482-9. [Crossref] [PubMed]
- Kohler C, Barekati Z, Radpour R, et al. Cell-free DNA in the circulation as a potential cancer biomarker. Anticancer Res 2011;31:2623-8. [PubMed]
- Krysko DV, Vanden Berghe T, D'Herde K, et al. Apoptosis and necrosis: detection, discrimination and phagocytosis. Methods 2008;44:205-21. [Crossref] [PubMed]
- Ulivi P, Mercatali L, Zoli W, et al. Serum free DNA and COX-2 mRNA expression in peripheral blood for lung cancer detection. Thorax 2008;63:843-4. [Crossref] [PubMed]
- van der Drift MA, Hol BE, Klaassen CH, et al. Circulating DNA is a non-invasive prognostic factor for survival in non-small cell lung cancer. Lung Cancer 2010;68:283-7. [Crossref] [PubMed]
- Ulivi P, Mercatali L, Casoni GL, et al. Multiple marker detection in peripheral blood for NSCLC diagnosis. PLoS One 2013;8:e57401. [Crossref] [PubMed]
- Chiappetta C, Anile M, Leopizzi M, et al. Use of a new generation of capillary electrophoresis to quantify circulating free DNA in non-small cell lung cancer. Clin Chim Acta 2013;425:93-6. [Crossref] [PubMed]
- Catarino R, Coelho A, Araújo A, et al. Circulating DNA: diagnostic tool and predictive marker for overall survival of NSCLC patients. PLoS One 2012;7:e38559. [Crossref] [PubMed]
- Kumar S, Guleria R, Singh V, et al. Efficacy of circulating plasma DNA as a diagnostic tool for advanced non-small cell lung cancer and its predictive utility for survival and response to chemotherapy. Lung Cancer 2010;70:211-7. [Crossref] [PubMed]
- Soliman SE, Alhanafy AM, Habib MSE, et al. Serum circulating cell free DNA as potential diagnostic and prognostic biomarker in non small cell lung cancer. Biochem Biophys Rep 2018;15:45-51. [Crossref] [PubMed]
- Szpechcinski A, Chorostowska-Wynimko J, Struniawski R, et al. Cell-free DNA levels in plasma of patients with non-small-cell lung cancer and inflammatory lung disease. Br J Cancer 2015;113:476-83. [Crossref] [PubMed]
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. [Crossref] [PubMed]
- Crowley E, Di Nicolantonio F, Loupakis F, et al. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol 2013;10:472-84. [Crossref] [PubMed]
- Li C, Yin Y, Liu X, et al. Non-small cell lung cancer associated microRNA expression signature: integrated bioinformatics analysis, validation and clinical significance. Oncotarget 2017;8:24564-78. [Crossref] [PubMed]
(English Language Editor: A. Kassem)