
EHD2 inhibits the invasive ability of lung adenocarcinoma and improves the prognosis of patients
Introduction
Lung cancer is the most lethal malignant tumor worldwide with the highest mortality rate (1). Non-small cell lung cancer (NSCLC) accounts for approximately 80–85% of lung cancers, and lung adenocarcinoma (LUAD) has become the most prevalent subtype (2). At present, radiotherapy is the most important non-surgical treatment for LUAD (3). Although radiotherapy technology has advanced significantly, the survival rate has plateaued (4). Unfortunately, radiotherapy resistance is one of the main reasons for the failure of LUAD treatment, and its molecular mechanism has not been fully elucidated.
The curative effect of radiotherapy is affected by factors such as epithelial-mesenchymal transition (EMT), tumor inherent radiation resistance, and the tumor microenvironment (TME) (5-7). E-cadherin forms vesicles and regulates adhesion junctions through endocytosis, and the down-regulation of E-cadherin expression is the key event of EMT (8,9). Meanwhile, specific modules such as the EH (Eps15 homology) domain regulate the entire endocytosis process (10). The EH domain contains protein 2 (EHD2), a member of EHD protein family, which is critical for nucleotide-dependent membrane remodeling and is involved in membrane transport between cell and plasma membranes (11,12). EHD2 is a mechanotransducing ATPase localized in caveolae invaginations at the plasma membrane. Its overexpression leads to suppression of internalization, and it may inhibit the migration and invasion of liver cancer by interacting with E-cadherin to improve prognosis (13). Research has shown that patients with colorectal cancer (CRC) who have EHD2 up-regulation experience longer overall survival (OS). Overexpressing EHD2 was shown to inhibit CRC cell migration and to increase the invasion, apoptosis, and cell cycle arrest (14,15). In addition, EHD2 has been implicated in glioma and esophageal squamous cell cancer as a tumor suppressor gene (16,17); these studies showed that EHD2 may be involved in tumorigenesis and development. Nonetheless, the role of EHD2 in LUAD is unknown.
In the present study, using bioinformatics analysis, we determined that EHD2 is critical in LUAD. Further experiments have shown that EHD2 can suppress LUAD migration and invasion by preventing EMT process. In addition, we analyzed the association between EHD2 level and prognosis in LUAD, suggesting that EHD2 may become a biomarker for predicting LUAD prognosis. We present the following article in accordance with the TRIPOD reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-842/rc).
Methods
Data acquisition
We obtained a dataset (535 LUAD and 59 normal samples) from The Cancer Genome Atlas (TCGA; https://portal.gdc.cancer.gov/) LUAD database. Meanwhile, 170 LUAD tissues were provided by the Affiliated Tumor Hospital of Nantong University in China. From 2012 to 2013, all patients received surgical resection. The average follow-up time was 70 months. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of the Affiliated Tumor Hospital of Nantong University (No. 2016-094). All participants provided written informed consent.
Biological information analysis
To detect EHD2 expression in LUAD tissues, we utilized the Tumor Immune Estimation Resource (TIMER) and Gene Expression Profiling Interactive Analysis (GEPIA) to construct box plots. At the same time, GEPIA was employed for analyzing the link between EHD2 and E-Cadherin. The correlation coefficient was determined by the Spearman method. Kaplan-Meier Plotter was deployed for determining the link between EHD2 and OS. Further, R software (The R Foundation for Statistical Computing, Vienna, Austria) was used to perform correlation testing on genes to find genes that have a co-expression relationship with the target gene. The selection conditions were (|cor|>0.4 & P<0.05). We found genes that have a co-expression relationship with the target gene, and used the pheatmap package to draw heat maps for the 20 most significant positive and negative correlations. We utilized the clusterProfiler package to conduct Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis on differential genes (P<0.05), producing a bubble chart.
Gene set enrichment analysis (GSEA)
GSEA was undertaken to identify genes with significant differences based on the expression of EHD2. The arrangement was set to 1,000. We then performed evaluation of the enriched KEGG pathway to determine the P value and standardized enrichment score.
Immunohistochemistry (IHC) and evaluation of immunostaining
The tissues were fixed in neutral buffered formalin (10%). After the tissues had been embedded tissues in paraffin, they were cut into sections. Antigen activity restoration was accomplished by boiling the sections in citrate buffer (10 mmol/L, pH 6.0) for 3 minutes in an autoclave after they had been deparaffinized in xylene and rehydrated with graded alcohol. A 3% hydrogen peroxide was employed to quench endogenous peroxidase activity. The antibodies employed for immunoassays included: anti-EHD2 (1:100 dilution, ab222888, Abcam, Cambridge, MA, USA). Immunostaining was undertaken utilizing avidin-biotin-peroxidase complex method; meanwhile chromogenic reagent diaminobenzidine was used to observe the antigen-antibody reaction. The negative control slides were treated with phosphate-buffered saline (PBS). Without knowing the clinical and pathological parameters of LUAD patients, a blind method was used to evaluate all immunostained sections. To score EHD2 staining, a semi-quantitative immune response scoring (IRS) system was implemented, which included distribution area and staining intensity. The immunostaining intensity score was 0 to 3 points (0, 1, 2, and 3 corresponded to no response, weak response, mild response, and strong response, respectively) and they were 1 (0–25%), 2 (26–50%), 3 (51–75%), and 4 (76–100%). A final score could be attained by multiplying the intensity score by the proportional score. The results of 0–4 and 5–12 indicated low and high, respectively. The above evaluation process was carried out by 2 independent pathologists using a multi-head microscope, and the 2 pathologists collaborated to reach a consensus.
Western blot analysis
To extract proteins, we utilized radioimmunoprecipitation assay (RIPA) buffer (Beyotime, China, Shanghai) involving protease inhibitor cocktail as well as phosphatase inhibitor (Bimake, Houston, TX, USA), and to identify total protein concentrations, we employed Bio-Rad protein assay (Bio-Rad, Hercules, CA, USA). After polyacrylamide gel electrophoresis (PAGE), the proteins were transferred to polyvinylidene fluoride (PVDF) membrane before being incubated with a specific primary antibody overnight at 4 ℃. Anti-EHD2 (ab222888), anti-Vimentin (ab92547), and anti-GAPDH (ab8245) were supplied by Abcam (Cambridge, MA, USA). Both anti-E-cadherin [14472] as well as anti-N-cadherin [13116] were provided by Cell Signaling Technology (Boston, MA, USA). Both goat anti-rabbit IgG-HRP (abs20040ss) as well as goat anti-mouse IgG-HRP (abs20039ss) were supplied by AiBiXin Biotechnology Co., Ltd. (Absin, China). The PVDF membrane was incubated with the secondary antibody for 1 hour at room temperature. The membrane was observed using ChemiDoc MP Imaging System from Bio-Rad, USA. We used ImageJ (National Institutes of Health, Bethesda, MD, USA) to analyze band intensity.
Cell culture and cell transfection
The cell lines A549, NCI-H1299, NCI-H1650, NCI-H1975, and NCI-H4006 were provided by Shanghai Institute of Cell Biology, and each cell line was authenticated. The above cell line was stored in Roswell Park Memorial Institute (RPMI)-1640 supplemented with 10% fetal bovine serum (FBS) and cultured at 37 ℃ in 5% CO2. We used a lentiviral vector supplied by Shanghai Heyuan (Shanghai, China) to establish stable transfected cell lines. Then, we added 2 mg/mL puromycin to select cells. Finally, we used western blot to evaluate the transfection efficiency. The EHD2 gene target sequences and short hairpin RNA (shRNA) sequences of EHD2 are shown in Table 1.
Table 1
| Gene | Sequences |
|---|---|
| EHD2 | |
| Forward primer (5'-3') | 5'-CGGAATTCCATGTTCAGCTGGCTG-3' |
| Reverse primer (5'-3') | 5'-CGGGATCCCTCGGCGGAGCCCTT-3' |
| shRNA | |
| EHD2-shRNA1 | 5'-CTCCCTAATCAGGTCCTGGAGAG-3' |
| EHD2-shRNA2 | 5'-CTGCACGCACACCCCTGCTCCGG-3' |
| EHD2-shRNA3 | 5'-AAGAAAGAGATGCCCACGGTGTT-3' |
PCR, polymerase chain reaction; EHD2, EH domain contains protein 2; shRNA, short hairpin RNA.
Phagokinetic motility assays
After adding 1 mL of PBS (containing 20 µg of fibronectin) to each well of the 6-well plate, the wells were then subjected to 2 hours incubation at 37 ℃ in 5% CO2. After washing with PBS, 86 µL of microspheres was added to 30 mL serum-free medium, gently pipetting to mix and homogenize, after which 2.5 mL of microglobulin-containing medium was added to each well. These were then incubated at 37 ℃ in 5% CO2 for 1 hour, the cells were resuspended in a medium mixed with 0.05% FBS, and then 1,500 cells were poured into in each well and incubated for 18 hours. Finally, we observed the cell migration trajectory under the microscope and recorded the length of the movement.
Transwell migration and invasion assays
The cell migration and invasion tests were undertaken by means of 24-well transwell plate (pore size 8 µm; Costar, Corning, NY, USA). Then, the upper chamber with a serum-free medium was charged with 5×104 cells, and the lower chamber was charged with a medium mixed with 10% FBS. Following 1-day incubation at 37 ℃ in 5% CO2 incubator, the cells in upper chamber were stained with crystal violet. The average of cell counts observed in 5 random fields was utilized for calculating migrating cell number. Before adding the cells to the upper chamber, 50 µL of diluted Matrigel [Becton, Dickinson, and Co. (BD) Biosciences, Franklin Lakes, NJ, USA] was added.
Statistical analysis
All data were analyzed using the software SPSS 24.0 (IBM Corp., Armonk, NY, USA) and R 3.6.2 (The R Foundation for Statistical Computing, Vienna, Austria). To analyze the results of IHC staining and clinical parameters, the Pearson’s chi-square test was employed. We defined OS as the time to death from any cause. The survival curve was determined using Kaplan-Meier approach, and the analysis was implemented utilizing log-rank test. To assess the associations between the EHD2 expression and OS outcomes, hazard ratios (HRs) with 95% confidence intervals (CIs) were assessed by using univariate and multivariate Cox regression analyses. Adjustment variables included age, gender, smoking history, pathologic stage, T stage, and lymph node metastasis. Statistical significance was indicated by a 2-sided P value <0.05.
Results
Bioinformatics analysis of LUAD revealed that EHD2 could be a biomarker
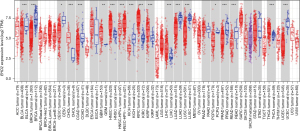
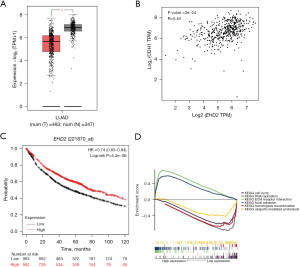
The TIMER database was employed to analyze EHD2 expression in different cancer types. The expression of EHD2 was down-regulated in most cancers and reached the highest value in adjacent tissues of LUAD (Figure 1). We also observed in the GEPIA database that EHD2 was significantly down-regulated in LUAD (Figure 2A), and it was significantly positively correlated with CDH1 expression (Figure 2B). Then, the Kaplan-Meier plotter indicated that down-regulating EHD2 expression was linked to poor prognosis in LUAD patients (Figure 2C). The GSEA analysis demonstrated that EHD2 may contribute to cell cycle progression, DNA replication, extracellular matrix (ECM) receptor interaction, focal adhesion, and homologous recombination (Figure 2D).
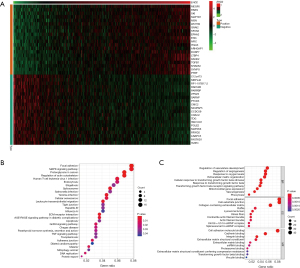
In addition, co-expression analysis was performed in TCGA-LUAD samples; co-expression was considered as |cor|>0.4, P<0.05 (Figure 3A). The GO and KEGG analyses revealed that EHD2 was associated with focal adhesion, tight junction, cadherin binding, DNA replication, and adherens junction (Figure 3B,3C). These findings cumulatively implied that EHD2 contributes to the EMT process of NSCLC and regulates radiosensitivity.
EHD2 expression and its link to clinicopathologic variables in LUAD cancer
To ascertain the role of EHD2 in LUAD, after preparing 4 LUAD tissue samples, we used western blot analysis to detect EHD2 expression. As illustrated in Figure 4A, LUAD tissues exhibited significantly lower EHD2 expression than adjacent normal tissues. To further determine EHD2 protein expression, we used IHC analysis to detect 170 tissue samples from LUAD patients. The EHD2 immune response was mainly located on the cell membrane (Figure 4B,4C). The IHC findings showed that EHD2 exhibited high expression in tissues without lymph node metastasis (Figure 4B). Nevertheless, tissues with lymph node metastasis had lower EHD2 expression (Figure 4C). Low EHD2 expression was intimately linked to advanced tumor-node-metastasis (TNM) stage (P=0.034) or lymph node metastasis (P=0.039) in LUAD (Table 2).
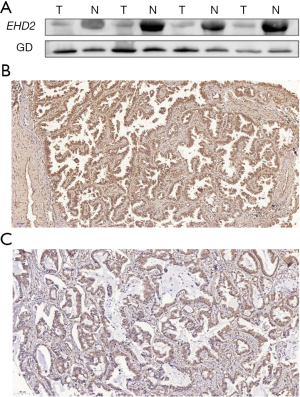
Table 2
| Variables | Patients | EHD2 low, n (%) | EHD2 high, n (%) | P value |
|---|---|---|---|---|
| Gender | 0.518 | |||
| Female | 69 | 30 (43.5) | 39 (56.5) | |
| Male | 101 | 49 (48.5) | 52 (51.5) | |
| Age (years) | 0.398 | |||
| <65 | 34 | 18 (52.9) | 16 (47.1) | |
| ≥65 | 136 | 61 (44.9) | 75 (55.1) | |
| Smoking history | 0.780 | |||
| No | 135 | 62 (45.9) | 73 (54.1) | |
| Yes | 35 | 17 (48.6) | 18 (51.4) | |
| Pathologic stage | 0.034* | |||
| I–II | 117 | 48 (41.0) | 69 (59.0) | |
| III–IV | 53 | 31 (58.5) | 22 (41.5) | |
| T stage | 0.868 | |||
| T1 and T2 | 145 | 67 (48.9) | 78 (51.1) | |
| T3 and T4 | 25 | 12 (72.7) | 13 (27.3) | |
| Lymph node metastasis | 0.039* | |||
| No | 94 | 37 (39.4) | 57 (60.6) | |
| Yes | 76 | 42 (55.3) | 34 (44.7) | |
*, P<0.05. EHD2, EH domain contains protein 2.
Prognostic significance of EHD2 expression
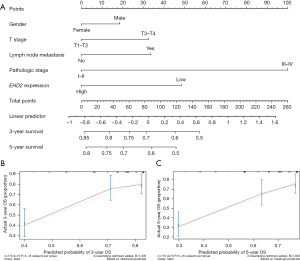
For univariate analysis, the Kaplan-Meier survival curve revealed that, except for patients without adjuvant radiotherapy, low EHD2 expression in other groups was significantly associated with low survival (Figure 5A-5C). The Cox proportional hazards regression model confirmed that independent prognostic factors for LUAD patients included EHD2 expression and TNM stage (Table 3). The nomogram which was established on the basis of the Fine and Gray models is displayed in Figure 6A. The calibration chart showed that these points were very close to the blue line, indicating a high degree of agreement between the predicted and actual survival events (Figure 6B,6C).

Table 3
| Variables | Univariate | Multivariate | |||
|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | ||
| Gender | 1.176 (0.733–1.884) | 0.502 | – | – | |
| Age (years) | 1.080 (0.603–1.934) | 0.796 | – | – | |
| Smoking history | 1.151 (0.661–2.004) | 0.619 | – | – | |
| Pathologic stage | 3.367 (2.120–5.346) | <0.01* | 3.284 (2.065–5.223) | <0.01* | |
| T stage | 2.009 (1.137–3.551) | 0.016* | – | – | |
| Lymph node metastasis | 2.092 (1.311–3.337) | <0.01* | – | – | |
| EHD2 expression | 0.572 (0.360–0.908) | 0.018* | 0.605 (0.381–0.963) | 0.034* | |
*, P<0.05. OS, overall survival; HR, hazard ratio; CI, confidence interval; EHD2, EH domain contains protein 2.
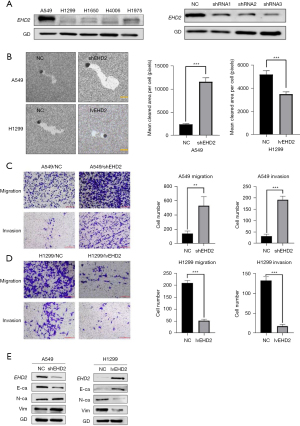
EHD2 suppresses EMT in LUAD cells
The expression of EHD2 in LUAD cell lines (A549, H1299, H1650, H1975, and H4006) was explored, demonstrating higher expression in A549 cells than in H1299, H1650, H1975, and H4006 cells (Figure 7A). Then, we used lentivirus-mediated infection to knockdown and overexpress EHD2 in the cell lines H1299 and A549. We used shRNA3 (EHD2 shRNA) in subsequent experiments because it was the most effective at knocking down EHD2 (Figure 7B). This comprehensive analysis indicated a link between EHD2 expression and lymph node metastasis. Therefore, we inferred that EHD2 contributes to migration and invasion inhibition. Transwell assays and phagocytic motility assays revealed that EHD2 knockdown markedly contributed to an increase in migration and invasion in A549 cells. By contrast, EHD2 overexpression substantially suppressed H1299 cell migration and invasion (Figure 7C,7D). Western blot analysis was executed to identify EMT markers, including E-cadherin, N-cadherin, and vimentin. The results indicated decreased E-cadherin expression in A549 cells with EHD2 knockdown, as well as increased N-cadherin and vimentin expressions. By contrast, E-cadherin expression was increased in H1299 cells, whereas N-cadherin and vimentin expressions were decreased in response to EHD2 overexpression (Figure 7E). These findings demonstrated that EHD2 suppressed EMT of LUAD.
Discussion
Lung cancer is one of the world’s most dangerous malignant tumors, posing a threat to human health and life. In terms of lung cancer cases, NSCLC is the most prevalent, accounting for up to 90% of all cases, and LUAD has become the prevalent type of NSCLC. Radiotherapy is one of the traditional clinical treatment options for NSCLC, but due to poor targeting and side effects, the prognosis of patients has been poor (18). Therefore, finding novel therapeutic targets has important therapeutic significance.
EHD2 is a member of EHD protein family with the lowest conservation degree, which is responsible for the membrane trafficking between the endosomes and plasma membrane (19,20). In muscle cells, EHD2 has been shown to be involved in membrane resealing/fusion, which can regulate actin function in several cell structures (20). These associations may change in normal cells adjacent to the tumor or in malignant cells, thereby promoting invasion, metastasis, and colonization (21). Previous studies have shown that EHD2 may serve as a prognostic marker in breast cancer, papillary thyroid carcinoma, esophageal squamous cell carcinoma, and CRC (15,17,21,22). Furthermore, Shen et al. reported that low EHD2 levels were associated with enhanced proliferation, migration, and invasion of triple negative breast cancer (TNBC) cells, and good prognosis in the highly proliferative TNBC subtype (23). In line with previous studies, we found that EHD2 was down regulated in LUAD tissues and correlated with poor prognosis through comprehensive bioinformatics analysis. Notably, while EHD2 expression was high in tissues free of lymph node metastasis in the present study, it was low in lymph node metastasis tissues. The decreased expression of EHD2 was associated with advanced TNM stage or lymph node metastasis. Furthermore, the multivariate Cox analysis showed that EHD2 expression levels were independent risk factors for survival following radical resection. These findings provide, for the first time, evidence that EHD2 may be a useful prognostic biomarker for LUAD.
EHD2 is implicated in regulating cell membrane transport, and is related to signal transduction regulation, as well as actin cytoskeleton and transcriptional regulation of endocytic pathway (11,24,25). The current research sought to explore the role of EHD2 in LUAD and its underlying molecular mechanism. The GSEA analysis showed that EHD2 may contribute to cell cycle progression, DNA replication, ECM receptor interaction, focal adhesion, and homologous recombination. In vitro, the present results showed that EHD2 knockdown markedly contributed to an increase in migration and invasion in LUAD cells and EHD2 overexpression substantially suppressed migration and invasion in LUAD cells. However, the present study did not assess the effects of EHD2 on proliferation or apoptosis in LUAD. By contrast, Liu et al. demonstrated that EHD2 overexpression enhanced the proliferation, invasion, and migration but inhibited the apoptosis of clear cell renal cell carcinoma (ccRCC) cells, while EHD2 interference showed opposite functions. Interference of EHD2 can inhibit the development of ccRCC by inhibiting the cellular proliferation, invasion, and migration (26). In addition, EHD2 overexpression inhibits colon cancer cell proliferation, but enhances cell apoptosis and cell cycle arrest (15). Regarding the role of EHD2 on proliferation or apoptosis in LUAD needs to be addressed in future studies.
In the present study, we also used western blot analysis to identify EMT markers, and the results showed that EHD2 increased E-cadherin expression, whereas it reduced the expressions of N-cadherin and vimentin. Previous studies have indicated that EHD2 knockdown promotes EMT, while EHD2 overexpression inhibits EMT in osteosarcoma and liver cancer (13,27). Several studies have demonstrated that EMT promotes early epithelial cancer cell diffusion and is an essential parameter in epithelial cancer invasion and metastasis (28,29). In addition, cancer cells undergoing EMT tend to be resistant to various anti-cancer treatments (30-32). To our knowledge, EHD2 regulates actin recombination to promote endocytosis by controlling Rac1 activity in tumorigenesis. Rac1 regulates the maintenance of cell polarity and cell migration by regulating GTPase activity and cytoskeleton rearrangement. It is an important signal transduction and polarity regulator in cells (10,15). This regulation of activity may significantly affect EMT, which is closely related to tumor cell invasion and distant metastasis (33). Furthermore, E-cadherin, as a calcium dependent cell adhesion molecule, can mediate the adhesion of allogeneic cells (34). The abnormal expression of E-cadherin is related to tumor differentiation, metastasis, invasion, and prognosis (35). With the decrease or disorder of E-cadherin expression, the ability of tumor invasion and metastasis will be enhanced (36,37). Given the above findings, EHD2 may have the potential to serve as therapeutic target for LUAD.
In conclusion, our study demonstrated that EHD2 reduces LUAD cell migration and invasion by preventing EMT. The expression level of EHD2 can be used as an independent prognostic factor for the survival of LUAD.
Acknowledgments
We thank Song He and Xinghua Zhu from the Department of Pathology of Nantong Tumor Hospital for their technical support.
Funding: This study was funded by the Scientific Research Project of Nantong Health Committee (No. MB2020016), and the Jiangsu Province’s Hospital Association (No. JSYGY-3-2019-381).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-842/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-842/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-842/coif). The authors have no conflicts of interest to declare.
Ethics Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of the Affiliated Tumor Hospital of Nantong University (No. 2016-094). All participants provided written informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. [Crossref] [PubMed]
- Bade BC, Dela Cruz CS. Lung Cancer 2020: Epidemiology, Etiology, and Prevention. Clin Chest Med 2020;41:1-24. [Crossref] [PubMed]
- Dong B, Wang J, Zhu X, et al. Comparison of the outcomes of stereotactic body radiotherapy versus surgical treatment for elderly (≥70) patients with early-stage non-small cell lung cancer after propensity score matching. Radiat Oncol 2019;14:195. [Crossref] [PubMed]
- Haseltine JM, Rimner A, Shepherd AF, et al. Delivering safe and effective stereotactic body radiation therapy for patients with centrally located early stage non-small cell lung cancer. Chin Clin Oncol 2020;9:39. [Crossref] [PubMed]
- Yin H, Wang X, Zhang X, et al. UBE2T promotes radiation resistance in non-small cell lung cancer via inducing epithelial-mesenchymal transition and the ubiquitination-mediated FOXO1 degradation. Cancer Lett 2020;494:121-31. [Crossref] [PubMed]
- Assani G, Zhou Y. Effect of modulation of epithelial-mesenchymal transition regulators Snail1 and Snail2 on cancer cell radiosensitivity by targeting of the cell cycle, cell apoptosis and cell migration/invasion. Oncol Lett 2019;17:23-30. [PubMed]
- Shintani Y, Okimura A, Sato K, et al. Epithelial to mesenchymal transition is a determinant of sensitivity to chemoradiotherapy in non-small cell lung cancer. Ann Thorac Surg 2011;92:1794-804; discussion 1804. [Crossref] [PubMed]
- Sommariva M, Gagliano N. E-Cadherin in Pancreatic Ductal Adenocarcinoma: A Multifaceted Actor during EMT. Cells 2020;9:1040. [Crossref] [PubMed]
- Akhtar N, Hotchin NA. RAC1 regulates adherens junctions through endocytosis of E-cadherin. Mol Biol Cell 2001;12:847-62. [Crossref] [PubMed]
- Benjamin S, Weidberg H, Rapaport D, et al. EHD2 mediates trafficking from the plasma membrane by modulating Rac1 activity. Biochem J 2011;439:433-42. [Crossref] [PubMed]
- Naslavsky N, Caplan S. EHD proteins: key conductors of endocytic transport. Trends Cell Biol 2011;21:122-31. [Crossref] [PubMed]
- Stoeber M, Stoeck IK, Hänni C, et al. Oligomers of the ATPase EHD2 confine caveolae to the plasma membrane through association with actin. EMBO J 2012;31:2350-64. [Crossref] [PubMed]
- Liu J, Ni W, Qu L, et al. Decreased Expression of EHD2 Promotes Tumor Metastasis and Indicates Poor Prognosis in Hepatocellular Carcinoma. Dig Dis Sci 2016;61:2554-67. [Crossref] [PubMed]
- Li J, Yang J, Hua L, et al. Ese-3 contributes to colon cancer progression by downregulating EHD2 and transactivating INPP4B. Am J Cancer Res 2021;11:92-107. [PubMed]
- Guan C, Lu C, Xiao M, et al. EHD2 Overexpression Suppresses the Proliferation, Migration, and Invasion in Human Colon Cancer. Cancer Invest 2021;39:297-309. [Crossref] [PubMed]
- Smith JS, Tachibana I, Pohl U, et al. A transcript map of the chromosome 19q-arm glioma tumor suppressor region. Genomics 2000;64:44-50. [Crossref] [PubMed]
- Li M, Yang X, Zhang J, et al. Effects of EHD2 interference on migration of esophageal squamous cell carcinoma. Med Oncol 2013;30:396. [Crossref] [PubMed]
- Buddharaju LNR, Ganti AK. Immunotherapy in lung cancer: the chemotherapy conundrum. Chin Clin Oncol 2020;9:59. [Crossref] [PubMed]
- Shah C, Hegde BG, Morén B, et al. Structural insights into membrane interaction and caveolar targeting of dynamin-like EHD2. Structure 2014;22:409-20. [Crossref] [PubMed]
- Morén B, Shah C, Howes MT, et al. EHD2 regulates caveolar dynamics via ATP-driven targeting and oligomerization. Mol Biol Cell 2012;23:1316-29. [Crossref] [PubMed]
- Yang X, Ren H, Yao L, et al. Role of EHD2 in migration and invasion of human breast cancer cells. Tumour Biol 2015;36:3717-26. [Crossref] [PubMed]
- Kim Y, Kim MH, Jeon S, et al. Prognostic implication of histological features associated with EHD2 expression in papillary thyroid carcinoma. PLoS One 2017;12:e0174737. [Crossref] [PubMed]
- Shen WW, Bièche I, Fuhrmann L, et al. EHD2 is a Predictive Biomarker of Chemotherapy Efficacy in Triple Negative Breast Carcinoma. Sci Rep 2020;10:7998. [Crossref] [PubMed]
- Naslavsky N, Caplan S. C-terminal EH-domain-containing proteins: consensus for a role in endocytic trafficking, EH? J Cell Sci 2005;118:4093-101. [Crossref] [PubMed]
- Marg A, Schoewel V, Timmel T, et al. Sarcolemmal repair is a slow process and includes EHD2. Traffic 2012;13:1286-94. [Crossref] [PubMed]
- Liu C, Liu S, Wang L, et al. Effect of EH domain containing protein 2 on the biological behavior of clear cell renal cell carcinoma. Hum Exp Toxicol 2019;38:927-37. [Crossref] [PubMed]
- Fan H, Liu T, Tian H, et al. TUSC8 inhibits the development of osteosarcoma by sponging miR-197-3p and targeting EHD2. Int J Mol Med 2020;46:1311-20. Erratum in: Int J Mol Med 2021;47(4):PMID: 32945345; PMCID: PMC7447318. [Crossref] [PubMed]
- Yang S, Liu Y, Li MY, et al. FOXP3 promotes tumor growth and metastasis by activating Wnt/β-catenin signaling pathway and EMT in non-small cell lung cancer. Mol Cancer 2017;16:124. [Crossref] [PubMed]
- Zhao YR, Wang JL, Xu C, et al. HEG1 indicates poor prognosis and promotes hepatocellular carcinoma invasion, metastasis, and EMT by activating Wnt/β-catenin signaling. Clin Sci (Lond) 2019;133:1645-62. [Crossref] [PubMed]
- Fischer KR, Durrans A, Lee S, et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 2015;527:472-6. [Crossref] [PubMed]
- Zheng X, Carstens JL, Kim J, et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 2015;527:525-30. [Crossref] [PubMed]
- Galle E, Thienpont B, Cappuyns S, et al. DNA methylation-driven EMT is a common mechanism of resistance to various therapeutic agents in cancer. Clin Epigenetics 2020;12:27. [Crossref] [PubMed]
- Pastushenko I, Blanpain C. EMT Transition States during Tumor Progression and Metastasis. Trends Cell Biol 2019;29:212-26. [Crossref] [PubMed]
- van Roy F, Berx G. The cell-cell adhesion molecule E-cadherin. Cell Mol Life Sci 2008;65:3756-88. [Crossref] [PubMed]
- Hirata K, Ajiki T, Okazaki T, et al. Frequent occurrence of abnormal E-cadherin/beta-catenin protein expression in advanced gallbladder cancers and its association with decreased apoptosis. Oncology 2006;71:102-10. [Crossref] [PubMed]
- Perez-Moreno M, Jamora C, Fuchs E. Sticky business: orchestrating cellular signals at adherens junctions. Cell 2003;112:535-48. [Crossref] [PubMed]
- Shi Y, Liu X, Sun Y, et al. Decreased expression and prognostic role of EHD2 in human breast carcinoma: correlation with E-cadherin. J Mol Histol 2015;46:221-31. [Crossref] [PubMed]
(English Language Editor: J. Jones)


