
Evaluation of a multiplex PCR kit for detection of 17 respiratory pathogens in hospitalized patients
Introduction
Pulmonary infections are the largest cause of human disease burden and carry high morbidity and mortality (1). Crude mortality was estimated at 20–60% for hospitalized patients with severe community-acquired pneumonia (2-4) and 30–70% for hospital-acquired pneumonia (5). Rapid identification of causative pathogens and prompt initiation of effective antimicrobial therapy are crucial for the prognosis of pneumonia patients, with significantly increased mortality reported among patients receiving delayed or ineffective treatment (6). In China, in-hospital mortality rates for diagnosed and undiagnosed patients are 21.7% and 25.9%, respectively (2).
Diagnosis of most bacterial and fungal infections depends on successful microbial or fungal culture, a process that takes about 48–72 h, prior to which patients can only receive empirical therapy. In addition, the low sensitivity of routine microbiology (approximately 44.2%) (7) that frequently limits patients to broad-spectrum treatment not only worsens prognosis but also increases the risk of adverse effects and promotes antibiotic resistance (8,9).
Multiplex polymerase chain reaction (PCR) offers rapid detection and precise identification of a large number of respiratory viruses by amplifying genomic fragments (10). A prior study reported a 30–50% increase in respiratory virus identification when using multiplex PCR as compared to antibody and culture methods (11). However, relatively few studies have investigated the use of multiplex PCR for the diagnosis of bacterial and fungal infections. Here, we designed and assessed a multiplex fluorescent PCR kit capable of detecting 17 respiratory pathogens (13 bacteria and 4 fungi) simultaneously in one PCR panel. We collected 452 respiratory and non-respiratory samples from hospitalized patients to evaluate the clinical performance of this kit and compared the results with those of routine microbiological tests. We present the following article in accordance with the STARD reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-544/rc).
Methods
Multiplex fluorescent PCR kit
The 17 respiratory pathogens targeted for detection by our multiplex PCR kit were as follows: Pseudomonas aeruginosa, Klebsiella pneumoniae, Acinetobacter baumannii, Staphylococcus aureus, Stenotrophomonas maltophilia, Streptococcus pneumoniae, group A streptococcus, Haemophilus influenzae, Enterococcus faecium, Enterococcus faecalis, Escherichia coli, Mycobacterium tuberculosis, atypical mycobacteria, Candida albicans, Mucor, Aspergillus and Penicillium marneffei. Target sequences corresponding to these pathogens were searched in the National Center of Biotechnology Information (NCBI); retrieved sequences were aligned using specialized software to select conserved regions with higher homology as targets for pathogen detection and thus ensure greater specificity. Corresponding primer and probe sequences were designed according to relevant pathogen conserved sequences (Table 1). The pre-designed multiplex fluorescent PCR kit panel included the primer and probe reaction mixture (4 µL) as well as nucleic acid amplification reaction solution (16 µL). The nucleic acid amplification reaction solution used as quantitative real time PCR (qPCR) reaction premix consisting of Tris (0.05–0.1 M), KCl (100–200 mM), MgCl2 (1–5 mM), dNTPs (0.1–0.8 mM) and Taqman (0.1–1 U/µL).
Table 1
| Pathogen | Forward primer (5'–3') | Reverse primer (5'–3') | Probe (5'–3') |
|---|---|---|---|
| Bacteria | |||
| Pseudomonas aeruginosa | TCGCAGACCAAGGACAAGCT | TTGCCCATCTCCTGTTCCA | TCTCCTCCGAGGTGAAGACCGCG |
| Klebsiella pneumoniae | TCCCCTTTGCCGTGAATAATC | GCGGCATACGCTGCTGTAT | CCCCGGTGGTCACCATTTCGG |
| Acinetobacter baumannii | TGCGACACAACTCGACGTTT | AATCTAGCACGACCTGACCATAGAC | TTTAAACCGATTGATTTGTCGCCGATCTTT |
| Staphylococcus aureus | TGATGGCTTTGAAGTAGTTTT | CACGATTCGAATAGTAAACATAA | TGCAGCAAGCCTTTTCTCTAAAATT |
| Stenotrophomonas maltophilia | CATGGCCAACGAAGAAAAGC | TGCAGGGTGTGGGTCACTT | TACGGCGTGCAGTTCCACCCG |
| Streptococcus pneumoniae | CCTCGTTGAAGCAATGGTGC | CCCAACAAGTGAATCACCAACA | TGGCATGGGCATGGTTGGTTTGGT |
| Group A streptococcus | CTCCTGGTGATCCCATACCAA | CTCCTGGTGATCCCATACCAA | TCCCACAAAGTCAGCACTGCTTAGACCA |
| Haemophilus influenzae | CCCAACAAGTGAATCACCAACA | TTCGCACATGAGCGTCAGTA | ACCGAAGGCGAAGGCAGCCCC |
| Enterococcus faecium | CAATGCTGCTTTGATACGAGTGT | AAGTTCTTGTCCGTGTTGACTTCA | CAGTGATCACGCCGTCTTTCAAAGGAA |
| Enterococcus faecalis | ACTTTGGTGTTGTTGAAGGTTTAATG | CCTTTAGGATGTGGTCCGTCTAA | CTATCCACGCTTACACAGGTGACCAAATGA |
| Escherichia coli | GCTGCTGTGGCGTCAAACT | GACCTACATGAGTGATTGCCTGAA | TTTTCACCAGGCGCAGACTTGCTGT |
| Mycobacterium tuberculosis | GCATCTGGCCACCTCGAT | GCCGCCAACTACGGTGTTTA | CCCTCACGGTTCAGGGTTAGCCACA |
| Atypical mycobacteria | TCCCGGGCCTTGTACACA | CCACTGGCTTCGGGTGTTA | CGCCCGTCACGTCATGAAAGTCG |
| Fungi | |||
| Candida albicans | ACCTGAAGTTTTACAATCAGCAACA | TGCTCGTAGCATTATCTATGCCTTA | TTACCAGCAGAATCAAAATGCACTTGACCA |
| Mucor | AGTACTTTGAAAAGAGAGTTAAACAG | GCTGATTAACTTCAAGTCAGTCT | TCGCAACCGACTCCATTAAGAACACCA |
| Aspergillus | GGCCGACAACAGCGTCAT | TCTGCTTGGCGGTGATGTAA | ATGTGAAATTGCCAAGAGGGAAGCATTTG |
| Penicillium marneffei | TGAAATTGTTCCTGCTCATGGT | CCACTCCCGTCGTAAATGTGT | TTCCTGGCATCCCTGTCAGCCATT |
| Internal reference | CTTCAGCATGGCGGTGTTT | CCGCGCAGAGCCTTCA | CAGATTTGGACCTGCGAGCGGG |
The plasmid used as a positive control for kit sensitivity evaluation was purchased from Shanghai Generay Biotech Co., Ltd. Target pathogen plasmids were mixed and diluted with Tris-EDTA to 100 copies/µL, 10 copies/µL, 2 copies/µL, 1 copy/µL, and 0.5 copies/µL, and then detected using the PCR kit. For sensitivity testing, a positive result was determined if the cycle threshold (Ct) value was ≤38 and the lowest copy number for which both tests were positive was considered as test sensitivity. Results revealed the minimum copy number of each target pathogen detected to have been 0.5 copies/µL and 1 copy/µL (Table 2).
Table 2
| Pathogen | 100 copies/μL | 10 copies/μL | 2 copies/μL | 1 copies/μL | 0.5 copies/μL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Value 1 (Ct) | Value 2 (Ct) | Value 1 (Ct) | Value 2 (Ct) | Value 1 (Ct) | Value 2 (Ct) | Value 1 (Ct) | Value 2 (Ct) | Value 1 (Ct) | Value 2 (Ct) | |||||
| Bacteria | ||||||||||||||
| Pseudomonas aeruginosa | 28.67 | 28.54 | 31.69 | 30.67 | 33.44 | 33.25 | 34.22 | 36.31 | 34.71 | 35.87 | ||||
| Klebsiella pneumoniae | 27.15 | 27.30 | 30.51 | 29.44 | 32.41 | 32.17 | 33.87 | 33.25 | 35.38 | 32.66 | ||||
| Acinetobacter baumannii | 29.95 | 29.88 | 33.27 | 32.99 | 36.06 | 35.59 | 35.57 | – | 37.12 | 36.71 | ||||
| Staphylococcus aureus | 28.89 | 29.01 | 32.84 | 32.04 | 34.37 | 34.90 | 36.73 | 35.43 | 36.13 | 36.17 | ||||
| Stenotrophomonas maltophilia | 29.11 | 29.12 | 32.23 | 32.14 | 34.86 | 35.65 | 37.62 | 35.76 | 37.67 | – | ||||
| Streptococcus pneumoniae | 29.37 | 29.47 | 33.16 | 30.06 | 34.54 | 35.46 | 35.24 | 37.19 | – | – | ||||
| Group A streptococcus | 27.59 | 27.60 | 31.32 | 30.83 | 33.17 | 33.28 | 35.00 | 33.79 | 35.71 | 35.50 | ||||
| Haemophilus influenzae | 30.64 | 30.91 | 35.48 | 35.06 | 38.78 | 35.12 | 34.82 | – | – | 36.95 | ||||
| Enterococcus faecium | 30.28 | 30.43 | 33.16 | 34.57 | 34.79 | 35.90 | – | 36.17 | 36.91 | – | ||||
| Enterococcus faecalis | 30.14 | 30.50 | 34.95 | 33.36 | 34.96 | 35.23 | 36.94 | 36.45 | – | – | ||||
| Escherichia coli | 28.97 | 29.25 | 32.22 | 32.37 | 36.10 | 34.39 | 35.91 | 34.88 | – | – | ||||
| Mycobacterium tuberculosis | 28.51 | 28.52 | 31.51 | 32.61 | 35.18 | 34.64 | 34.89 | 36.10 | – | 36.82 | ||||
| Atypical mycobacteria | 27.91 | 27.52 | 30.87 | 31.05 | 34.35 | 34.69 | 34.46 | 34.36 | 33.90 | 35.97 | ||||
| Fungi | ||||||||||||||
| Candida albicans | 26.350 | 28.848 | 30.651 | 33.690 | 34.477 | 36.970 | 36.164 | 34.976 | 36.517 | 36.377 | ||||
| Mucor | 27.431 | 27.457 | 30.505 | 31.309 | 35.603 | 33.693 | 35.324 | 35.225 | 35.571 | 36.694 | ||||
| Aspergillus | 25.937 | 25.928 | 27.874 | 31.236 | 32.483 | 32.921 | 33.645 | 34.419 | 34.821 | 34.613 | ||||
| Penicillium marneffei | 27.053 | 27.564 | 32.355 | 31.663 | 34.128 | 33.295 | 33.570 | 35.512 | 36.055 | 36.943 | ||||
PCR, polymerase chain reaction; Ct, cycle threshold value.
Samples
According to the number of samples required for positive validation and negative controls, a total of 483 samples (452 of them eligible) were discontinuously collected from infectious disease patients admitted to the Department of Pulmonary Medicine at Zhongshan Hospital, Fudan University between December 2020 and October 2021. Specimens of sufficient volume for two repeated PCR tests (>1 mL) and had at least one available test result on routine microbiological testing were included. The 452 eligible samples included 242 respiratory samples and 210 non-respiratory samples. Respiratory samples consisted of 206 sputum and 36 lower respiratory tract samples (endotracheal aspirates, bronchoalveolar lavage fluid and lung tissue). Non-respiratory samples consisted of 62 excrement (feces and urine) and 148 other body fluid and tissue samples (pleural effusion, ascitic fluid, cerebrospinal fluid, bile, pericardial fluid). After sample collection, approximately 1 mL of specimen was aliquoted per sample and stored at −80 ℃ for multiplex PCR testing within one week. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Medical Ethics Committee of Zhongshan Hospital, Fudan University (approval No. B2019-249R) and written informed consent was obtained from all patients.
Nucleic acid extraction
After homogenization, enzyme digestion or sample centrifugation, homogenous samples (300 µL) were transferred to a sample lysis plate and subjected to automated nucleic acid extraction using a nucleic acid extraction kit (Cat. #TQ-BG-001; BioGerm, Shanghai, China) and a BG-Abot-96 automated nucleic acid extraction system (BioGerm) according to manufacturer instructions.
Multiplex fluorescent PCR detection of respiratory pathogens
Nucleic acid extracted from all 452 specimens was subjected to multiplex amplification using our multiplex fluorescent PCR kit for respiratory pathogen detection. All PCR reactions were carried out using 5 µL of resuspended deoxyribonucleic acid (DNA), 4 µL of primer/probe mixture and 16 µL of nucleic acid amplification reaction solution. Optimal cycling conditions for nucleic acid amplification were initially 5 min at 95 ℃ followed by 40 cycles at 95 ℃ for 10 s and 55 ℃ for 40 s using an ABI 7500 (Invitrogen, Waltham, USA) device. It took a total of 80 min to complete pathogen detection. A positive result was defined by Ct values of ≤35 or 35–38 confirmed on repeat testing. Clinical information and routine microbiological tests results were not provided to researchers performing and evaluating multiplex PCR testing.
Routine microbiological result collection
All routine microbiological tests were performed according to requests of clinicians based on relevant standard operating procedures. Routine microbiological tests including bacterial and fungal smears and cultures, mycobacterial acid-fast smear and culture, and Aspergillus culture; data were collected from medical records upon conclusion of multiplex PCR testing. Clinical data and multiplex PCR test results were not available to performers of routine microbiological examinations.
Statistical analyses
Statistical analyses were performed using SPSS version 20 (IBM, Armonk, USA) and MedCalc version 20.027 (MedCalc Ltd. Ostend, Belgium). Statistical comparisons were analyzed using the chi-squared test. Sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) were analyzed for each PCR target considering routine microbiology as the reference standard (smears and cultures are the traditional diagnostic criteria for the 17 pathogens detected by our kit) (12).
Results
Routine microbiology results
Conventional bacterial smears and cultures were performed using 442 samples for detection of Pseudomonas aeruginosa, Klebsiella pneumoniae, Acinetobacter baumannii, Staphylococcus aureus, Stenotrophomonas maltophilia, Streptococcus pneumoniae, group A streptococcus, Haemophilus influenzae, Enterococcus faecium, Enterococcus faecalis and Escherichia coli (Table 3). Conventional fungal smears and cultures were performed on 405 samples to identify Candida albicans, Mucor, Aspergillus and Penicillium marneffei (Table 3). A total of 145 samples were evaluated using acid-fast smear and culture for M. tuberculosis. Aspergillus cultures were performed on 93 samples (Table 3). Presence of one or more pathogens was noted in 56.9% (257/452) of all samples; the positivity rate in respiratory and non-respiratory samples was 74.0% (179/242) and 37.1% (78/210), respectively.
Table 3
| Pathogen | Positive rate | ||
|---|---|---|---|
| All samples (n=452) | Respiratory samples (n=242) | Non-respiratory samples (n=210) | |
| Bacteria | |||
| Pseudomonas aeruginosa | 15.8% (70/442) | 22.8% (54/237) | 7.8% (16/205) |
| Klebsiella pneumoniae | 6.8% (30/442) | 8.4% (20/237) | 4.9% (10/205) |
| Acinetobacter baumannii | 19.7 (87/442) | 23.6% (56/237) | 15.1% (31/205) |
| Staphylococcus aureus | 5.4% (24/442) | 7.2% (17/237) | 3.4% (7/205) |
| Stenotrophomonas maltophilia | 3.6% (16/442) | 5.1% (12/237) | 2.0% (4/205) |
| Streptococcus pneumoniae | 0% (0/442) | 0% (0/237) | 0% (0/205) |
| Group A streptococcus | 0% (0/442) | 0% (0/237) | 0% (0/205) |
| Haemophilus influenzae | 1.4% (6/442) | 2.5% (6/237) | 0% (0/205) |
| Enterococcus faecium | 3.6% (16/442) | 1.7% (4/237) | 5.9% (12/205) |
| Enterococcus faecalis | 2.3% (10/442) | 1.3% (3/237) | 3.4% (7/205) |
| Escherichia coli | 2.0% (9/442) | 0.8% (2/237) | 3.4% (7/205) |
| Mycobacterium tuberculosis | 6.2% (9/145) | 8.6% (8/93) | 1.9% (1/52) |
| Atypical mycobacteria | 5.5% (8/145) | 8.6% (8/93) | 0% (0/52) |
| Fungi | |||
| Candida albicans | 11.9% (48/405) | 19.1% (43/225) | 2.8% (5/180) |
| Mucor | 0% (0/405) | 0% (0/225) | 0% (0/180) |
| Aspergillus | 6.5% (6/93) | 6.6% (6/91) | 0% (0/2) |
| Penicillium marneffei | 0% (0/405) | 0% (0/225) | 0% (0/180) |
Among the 442 samples studied using bacterial smear and culture, Acinetobacter baumannii and Pseudomonas aeruginosa were the most frequently noted pathogens among both respiratory and non-respiratory samples (Figure 1). Acinetobacter baumannii was found in 23.6% (56/237) of respiratory and 15.1% (31/205) of non-respiratory samples, while Pseudomonas aeruginosa was found in 22.8% (54/237) of respiratory and 7.8% (16/205) of non-respiratory samples (Table 3). Klebsiella pneumoniae was the third most frequently detected pathogen (8.3%) in respiratory samples (Figure 1A), while Enterococcus faecium was the third most frequently detected pathogen (5.9%) in non-respiratory samples (Figure 1B). Notably, neither Streptococcus pneumoniae nor group A streptococcus was noted among the 442 samples evaluated (Table 3; Figure 1). Similarly, Haemophilus influenzae and atypical mycobacteria were not detected among the 210 non-respiratory samples (Figure 1B). On fungal smear and culture, Candida albicans was found in 43 respiratory and five non-respiratory samples, while Aspergillus was found in six respiratory samples (Figure 1).
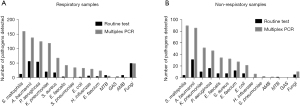
Multiplex PCR results
The overall positive rate for multiplex PCR detection exceeded that of routine microbiology (86.9%; 393/452); overall positive rates in respiratory and non-respiratory specimens were 94.2% (228/242) and 78.6% (165/210), respectively. Furthermore, the most frequently detected pathogens on multiplex PCR were Stenotrophomonas maltophilia in both respiratory (65.7%; 159/242) and non-respiratory (42.4%; 89/210) samples (Figure 1). In respiratory samples, the other most frequently detected pathogens were Acinetobacter baumannii (56.6%; 137/242), Pseudomonas aeruginosa (51.2%; 124/242), Klebsiella pneumoniae (48.8%; 118/242) and Staphylococcus aureus (27.7%; 67/242) (Figure 1A; Table 4). Among non-respiratory samples, the other four most frequently noted pathogens were Acinetobacter baumannii (40.5%; 85/210), Klebsiella pneumoniae (24.3%; 51/210), Pseudomonas aeruginosa (21.9%; 46/210) and Enterococcus faecalis (16.2%; 34/210) (Figure 1B; Table 4). Streptococcus pneumoniae was found in 36 samples using multiplex PCR but not routine microbiology (Tables 3,4).
Table 4
| Pathogen | Positive rate | ||
|---|---|---|---|
| All samples (n=452) | Respiratory samples (n=242) | Non-respiratory samples (n=210) | |
| Bacteria | |||
| Pseudomonas aeruginosa | 37.6% (170/452) | 51.2% (124/242) | 21.9% (46/210) |
| Klebsiella pneumoniae | 37.4% (169/452) | 48.8% (118/242) | 24.3% (51/210) |
| Acinetobacter baumannii | 49.1% (222/452) | 56.6% (137/242) | 40.5% (85/210) |
| Staphylococcus aureus | 21.9% (99/452) | 27.7% (67/242) | 15.2% (32/210) |
| Stenotrophomonas maltophilia | 54.9% (248/452) | 65.7% (159/242) | 42.4% (89/210) |
| Streptococcus pneumoniae | 8.0% (36/452) | 14.0% (34/242) | 1.0% (2/210) |
| Group A streptococcus | 0.9% (4/452) | 1.7% (4/242) | 0% (0/210) |
| Haemophilus influenzae | 7.7% (35/452) | 12.0% (29/242) | 2.9% (6/210) |
| Enterococcus faecium | 11.1% (50/452) | 9.5% (23/242) | 12.9% (27/210) |
| Enterococcus faecalis | 16.8% (76/452) | 17.4% (42/242) | 16.2% (34/210) |
| Escherichia coli | 11.7% (53/452) | 13.2% (32/242) | 10.0% (21/210) |
| Mycobacterium tuberculosis | 1.1% (5/452) | 2.1% (5/242) | 0% (0/210) |
| Atypical mycobacteria | 0.4% (2/452) | 0.4% (1/242) | 0.5% (1/210) |
| Fungi | |||
| Candida albicans | 11.5% (52/452) | 18.2% (44/242) | 3.8% (8/210) |
| Mucor | 0.2% (1/452) | 0% (0/242) | 0.5% (1/210) |
| Aspergillus | 0.9% (4/452) | 1.2% (3/242) | 0.5% (1/210) |
| Penicillium marneffei | 0.2% (1/452) | 0.4% (1/242) | 0% (0/210) |
PCR, polymerase chain reaction.
No atypical mycobacteria were detected in respiratory samples and no Mycobacterium tuberculosis or group A streptococcus were detected in non-respiratory samples (Table 4). Multiplex PCR results revealed 48 cases of fungi in respiratory samples, including 44 Candida albicans, 3 Aspergillus and 1 Penicillium marneffei, as well as 10 cases in non-respiratory samples, including 8 Candida albicans, 1 Mucor and 1 Aspergillus (Table 4).
Comparison of routine microbiology and multiplex PCR kit pathogen detection
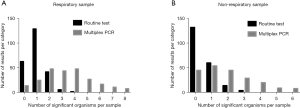
Not only was the overall positive rate of pathogen detection on multiplex PCR significantly higher than that of routine microbiology, but the rate of multiple organisms detection was also markedly greater. As shown in Figure 2, 155 respiratory (64.0%) and 66 non-respiratory (31.4%) samples were found to be positive for three or more pathogens on multiplex PCR, significantly greater than the eight respiratory (3.3%) and four non-respiratory (1.9%) samples evaluated using routine microbiology. Moreover, no specimens were positive for five or more pathogens on routine microbiology; multiplex PCR revealed 63 respiratory and 17 non-respiratory samples to be positive for five or more pathogens (Figure 2).
The overall proportion of agreement (concordant positive/all positive results on routine microbiology) among multiplex PCR and routine microbiology for detection and identification of the aforementioned bacteria was found to be 95.8%. The sensitivity of multiplex PCR was >90% for most target bacteria; NPVs were >99%, especially for Pseudomonas aeruginosa, Klebsiella pneumoniae, Acinetobacter baumannii, Stenotrophomonas maltophilia, Enterococcus faecium, Enterococcus faecalis and Escherichia coli (Table 5). Specificity and PPV were lower as multiplex PCR detected more pathogens per sample and revealed more positive samples as compared to routine microbiology (Table 5). Test result consistency (Table S1) also revealed that multiplex PCR had higher sensitivity for detection of most bacteria and thus positivity coincident rates were relatively low. Streptococcus pneumoniae and group A streptococcus were not detected on routine microbiology but were found in 36 and four samples, respectively, on multiplex PCR; as such their relevant specificities were high but sensitivities could not be calculated (Tables 3-5). However, lower sensitivity (33.3–59.6%) and higher specificity (93.9–100.0%) for detection of fungi and mycobacteria was noted using our multiplex PCR kit (Table 5). Although the number of fungi detected using multiplex PCR was close to that of routine microbiology, the concordant rate of positivity between the two methods was poor.
Table 5
| Pathogen | All samples (n=452) | Respiratory samples (n=242) | Non-respiratory samples (n=210) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | |||
| Bacteria | ||||||||||||||
| Pseudomonas aeruginosa | 97.1 | 72.8 | 40.2 | 99.3 | 98.1 | 61.7 | 43.1 | 99.1 | 93.8 | 83.6 | 32.6 | 99.4 | ||
| Klebsiella pneumoniae | 96.7 | 66.5 | 17.4 | 99.6 | 95.0 | 54.8 | 16.2 | 99.2 | 100.0 | 79.5 | 20.0 | 100.0 | ||
| Acinetobacter baumannii | 95.4 | 61.1 | 37.6 | 98.2 | 96.4 | 54.1 | 39.4 | 98.0 | 93.5 | 68.4 | 34.5 | 98.3 | ||
| Staphylococcus aureus | 87.5 | 81.6 | 21.4 | 99.1 | 88.2 | 76.8 | 22.7 | 98.8 | 85.7 | 86.9 | 18.8 | 99.4 | ||
| Stenotrophomonas maltophilia | 93.8 | 46.7 | 6.2 | 99.5 | 100.0 | 35.6 | 7.6 | 100.0 | 75.0 | 59.2 | 3.5 | 99.3 | ||
| Streptococcus pneumoniae | NA | 92.5 | 0 | 100.0 | NA | 86.9 | 0 | 100.0 | NA | 99.0 | 0 | 100.0 | ||
| Group A streptococcus | NA | 99.1 | 0 | 100.0 | NA | 98.3 | 0 | 100.0 | NA | 100.0 | 100.0 | 100.0 | ||
| Haemophilus influenzae | 83.3 | 93.1 | 14.3 | 99.8 | 83.3 | 89.6 | 17.2 | 99.5 | NA | 97.1 | 0 | 100.0 | ||
| Enterococcus faecium | 87.5 | 91.5 | 28.0 | 99.5 | 75.0 | 91.4 | 13.0 | 99.5 | 91.7 | 91.7 | 40.7 | 99.4 | ||
| Enterococcus faecalis | 90.0 | 84.5 | 11.8 | 99.7 | 100.0 | 83.3 | 7.1 | 100.0 | 85.7 | 85.9 | 17.6 | 99.4 | ||
| Escherichia coli | 100.0 | 89.8 | 17.0 | 100.0 | 100.0 | 87.2 | 6.3 | 100.0 | 100.0 | 92.9 | 33.3 | 100.0 | ||
| Mycobacterium tuberculosis | 33.3 | 100.0 | 100.0 | 95.8 | 25.0 | 98.8 | 66.7 | 93.3 | 0 | 100.0 | NA | 98.1 | ||
| Atypical mycobacteria | 0 | 100.0 | NA | 94.5 | 0 | 100.0 | NA | 91.4 | NA | 100.0 | 100.0 | 100.0 | ||
| Fungi | ||||||||||||||
| Candida albicans | 59.6 | 93.9 | 56.0 | 94.6 | 58.1 | 90.1 | 58.1 | 90.1 | 75.0 | 97.7 | 42.9 | 99.4 | ||
| Mucor | NA | 100.0 | 100.0 | 100.0 | NA | 100.0 | 100.0 | 100.0 | NA | 100.0 | 100.0 | 100.0 | ||
| Aspergillus | 50.0 | 100.0 | 100.0 | 96.7 | 50.0 | 100.0 | 100.0 | 96.6 | NA | 100.0 | 100.0 | 100.0 | ||
| Penicillium marneffei | 0 | 100.0 | NA | 99.3 | NA | 100.0 | 100.0 | 100.0 | NA | 100.0 | 100.0 | 100.0 | ||
PCR, polymerase chain reaction; PPV, positive predictive value; NPV, negative predictive value; NA, not available.
Discussion
In recent years, multiplex PCR has emerged as a simple, rapid and highly sensitive method for detection of respiratory pathogens (10,13). Here, we designed a multiplex fluorescent PCR kit for the identification of 17 respiratory pathogens simultaneously and evaluated its performance using 452 clinical samples obtained from inpatients. Sensitivity, specificity, PPV and NPV were analyzed for each PCR target considering routine microbiology as the gold standard.
Based on fluorescent PCR principles, we designed specific primers and Taqman probes for pathogen detection using a fluorescent PCR instrument. Our kit was designed to simultaneously detect 13 bacteria (Pseudomonas aeruginosa, Klebsiella pneumoniae, Acinetobacter baumannii, Staphylococcus aureus, Stenotrophomonas maltophilia, Streptococcus pneumoniae, group A streptococcus, Haemophilus influenzae, Enterococcus faecium, Enterococcus faecalis, Escherichia coli, Mycobacterium tuberculosis, and atypical mycobacteria), as well as four fungi (Candida albicans, Mucor, Aspergillus, and Penicillium marneffei) in one PCR panel. Compared with routine microbiology testing, which usually takes at least 48–72 h to obtain the culture result and requires higher sample quality, the multiplex PCR kit can complete the detection of 17 pathogens in only 80 min. Our kit is thus useful for the auxiliary diagnosis and epidemiological surveillance of respiratory pathogens.
The overall positive rate of pathogen detection (86.9% vs. 56.9%) and the detection rate of multiple organisms on multiplex PCR were both significantly higher than those of routine microbiology. Medical record review revealed detection of relevant pathogens on routine microbiology after significant illness progression; multiplex PCR was capable of detecting pathogens rapidly and with greater sensitivity. Clinical use of multiplex PCR thus offers a greater possibility of earlier diagnosis and treatment initiation.
In this study, clinical data analyzed were obtained from patients suffering nosocomial infections. In agreement with relevant prior literature (14), our routine microbiology findings revealed Acinetobacter baumannii, Pseudomonas aeruginosa and Klebsiella pneumoniae to have been the three predominant pathogens in respiratory samples, while Acinetobacter baumannii, Pseudomonas aeruginosa and Enterococcus faecium were found to have been the three predominant pathogens in non-respiratory samples. However, the pathogen most frequently detected by multiplex PCR in both respiratory and non-respiratory specimens was Stenotrophomonas maltophilia, and even it was found in more than half of respiratory specimens. As recent literature has reported the incidence of hospital-acquired Stenotrophomonas maltophilia infections to be on the increase (15), particularly among immunocompromised patients (16), its high rate of detection here warrants attention. Although Stenotrophomonas maltophilia is not highly virulent, it has emerged as an important nosocomial pathogen associated with a high mortality among patients suffering bacteremia that ranges from 14% to 69% (15,17,18). Inappropriate initial empirical antibiotic therapy is one of the risk factors for poor outcomes in the setting of Stenotrophomonas maltophilia pneumonia (18). Thus, the high sensitivity of multiplex PCR in Stenotrophomonas maltophilia identification is bound to assist clinicians in formulating more effective treatment regimens earlier and avoiding unreasonable antibiotic administration. Greater detection of Streptococcus pneumoniae on multiplex PCR is similarly important. Although one of the most common causes of pneumonia (19,20), the positive rate of Streptococcus pneumoniae detection on routine microbiological culture remains relatively low due to its fastidious nature (21,22). None of the 452 samples evaluated in this study were positive for Streptococcus pneumoniae on routine culture, but multiplex PCR successfully revealed Streptococcus pneumoniae in 36 samples. This advantage of multiplex PCR is bound to help clinicians more accurately establish diagnoses and perform relevant drug susceptibility testing.
Our results revealed that multiplex PCR has a high overall agreement with routine microbiology regarding the detection of bacteria commonly associated with pneumonia. Here, the detection sensitivity for most target bacteria was >90%, with NPVs >99%. One potential challenge of clinically implementing multiplex PCR use is that it detects additional bacteria not reported on routine microbiology (23). The number of bacteria detected by multiplex PCR is more than twice that detected using routine microbiology. Although this phenomenon underscores the high sensitivity of multiplex PCR, it remains necessary to distinguish possible causes of false positive results. First, some of the discordant results (i.e., multiplex PCR positive but culture-negative) found in this study might have been caused by administration of antimicrobial therapy prior to sampling (13). Because nucleic acids persist in vivo for some time, their presence does not necessarily signify ongoing infection. Another challenge posed by nucleic acid diagnostics for respiratory pathogen detection is the difficulty of distinguishing between colonization and infection (13,24). The presence of non-colonizing bacteria may certainly indicate an infectious etiology, but some respiratory pathogens such as Streptococcus pneumoniae and Candida albicans commonly colonize the upper respiratory tract and thus may present a diagnostic dilemma (25,26). It is thus important to establish a relationship between organism load and clinical context based on factors such as pathogen colonization characteristics to accurately determine etiology (27,28). In the further research plan, we will explore to distinguish infection from contamination or colonization by defining different-level cut-off values.
This study was not without its limitations. Due to insufficient relatively small number of samples, not enough cases of several uncommon pathogens were available for investigation. Thus, a larger-scale study is required to confirm our findings. Furthermore, since routine microbiology testing is not the true gold standard for pathogen detection, considering routine tests as the gold standard for comparison with multiplex PCR may obscure true multiplex PCR effectiveness. Future comparisons based on final diagnoses established by clinicians and documented patient outcomes are warranted.
Conclusions
In summary, our multiplex PCR kit designed to identify 17 respiratory pathogens possesses the advantages of high sensitivity and rapid turn-around. Thus, it is capable of aiding rapid identification of infectious etiologies. Importantly, the lower sensitivities of our kit regarding detection of mycobacteria and fungi require further study and enhancement. In addition, awareness of the clinical significance of potential false-positive results is critical when using our kit in practice. This, in turn, requires comprehensive judgment based on specific clinical manifestations and disease conditions of individual patients to accurately determine both etiology and treatment options (29). A large-scale, prospective study is required to confirm our findings and further explore the clinical effects of our multiplex PCR kit on patient management.
Acknowledgments
The researchers are grateful to all members of Prof. Song’s group and the Department of Laboratory Medicine of Zhongshan Hospital for their assistance.
Funding: This work was supported by the National Key R&D Plan (No. 2020YFC2003700), National Natural Science Foundation of China (Nos. 82130001, 82070045), Shanghai Sailing Program (No. 22YF1406100), Science and Technology Commission of Shanghai Municipality (Nos. 20Z11901000, 20DZ2261200, 20XD1401200), Shanghai Municipal Science and Technology Major Project, Clinical Research Plan of SHDC (No. SHDC2020CR5010-002) and Shanghai Municipal Key Clinical Specialty (No. shslczdzk02201).
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-544/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-544/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-544/coif). WG and YS have a patent of a composition, test kit for detecting respiratory pathogens and their application pending. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Medical Ethics Committee of Zhongshan Hospital, Fudan University (approval No. B2019-249R) and written informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lozano R, Naghavi M, Foreman K, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012;380:2095-128. [Crossref] [PubMed]
- Qu J, Zhang J, Chen Y, et al. Aetiology of severe community acquired pneumonia in adults identified by combined detection methods: a multi-centre prospective study in China. Emerg Microbes Infect 2022;11:556-66. [Crossref] [PubMed]
- Cilloniz C, Ferrer M, Liapikou A, et al. Acute respiratory distress syndrome in mechanically ventilated patients with community-acquired pneumonia. Eur Respir J 2018;51:1702215. [Crossref] [PubMed]
- Lenz H, Norby GO, Dahl V, et al. Five-year mortality in patients treated for severe community-acquired pneumonia – a retrospective study. Acta Anaesthesiol Scand 2017;61:418-26. [Crossref] [PubMed]
- Torres A, Niederman MS, Chastre J, et al. International ERS/ESICM/ESCMID/ALAT guidelines for the management of hospital-acquired pneumonia and ventilator-associated pneumonia: Guidelines for the management of hospital-acquired pneumonia (HAP)/ventilator-associated pneumonia (VAP) of the European Respiratory Society (ERS), European Society of Intensive Care Medicine (ESICM), European Society of Clinical Microbiology and Infectious Diseases (ESCMID) and Asociación Latinoamericana del Tórax (ALAT). Eur Respir J 2017;50:1700582. [Crossref] [PubMed]
- Piskin N, Aydemir H, Oztoprak N, et al. Inadequate treatment of ventilator-associated and hospital-acquired pneumonia: risk factors and impact on outcomes. BMC Infect Dis 2012;12:268. [Crossref] [PubMed]
- Enne VI, Aydin A, Baldan R, et al. Multicentre evaluation of two multiplex PCR platforms for the rapid microbiological investigation of nosocomial pneumonia in UK ICUs: the INHALE WP1 study. Thorax 2022; Epub ahead of print. [Crossref] [PubMed]
- Enne VI, Personne Y, Grgic L, et al. Aetiology of hospital-acquired pneumonia and trends in antimicrobial resistance. Curr Opin Pulm Med 2014;20:252-8. [Crossref] [PubMed]
- van Hecke O, Wang K, Lee JJ, et al. Implications of Antibiotic Resistance for Patients’ Recovery From Common Infections in the Community: A Systematic Review and Meta-analysis. Clin Infect Dis 2017;65:371-82. [Crossref] [PubMed]
- Huang HS, Tsai CL, Chang J, et al. Multiplex PCR system for the rapid diagnosis of respiratory virus infection: systematic review and meta-analysis. Clin Microbiol Infect 2018;24:1055-63. [Crossref] [PubMed]
- Babady NE. The FilmArray® respiratory panel: an automated, broadly multiplexed molecular test for the rapid and accurate detection of respiratory pathogens. Expert Rev Mol Diagn 2013;13:779-88. [Crossref] [PubMed]
- Cao B, Huang Y, She DY, et al. Diagnosis and treatment of community-acquired pneumonia in adults: 2016 clinical practice guidelines by the Chinese Thoracic Society, Chinese Medical Association. Clin Respir J 2018;12:1320-60. [Crossref] [PubMed]
- Lee SH, Ruan SY, Pan SC, et al. Performance of a multiplex PCR pneumonia panel for the identification of respiratory pathogens and the main determinants of resistance from the lower respiratory tract specimens of adult patients in intensive care units. J Microbiol Immunol Infect 2019;52:920-8. [Crossref] [PubMed]
- Kollef MH, Torres A, Shorr AF, et al. Nosocomial Infection. Crit Care Med 2021;49:169-87. [Crossref] [PubMed]
- Brooke JS. Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin Microbiol Rev 2012;25:2-41. [Crossref] [PubMed]
- Looney WJ, Narita M, Mühlemann K. Stenotrophomonas maltophilia: an emerging opportunist human pathogen. Lancet Infect Dis 2009;9:312-23. [Crossref] [PubMed]
- Zhu L, Wang L, Zhang Y, et al. Fatal hemorrhagic pneumonia in patients with hematologic diseases and Stenotrophomonas maltophilia bacteremia: a retrospective study. BMC Infect Dis 2021;21:723. [Crossref] [PubMed]
- Velázquez-Acosta C, Zarco-Márquez S, Jiménez-Andrade MC, et al. Stenotrophomonas maltophilia bacteremia and pneumonia at a tertiary-care oncology center: a review of 16 years. Support Care Cancer 2018;26:1953-60. [Crossref] [PubMed]
- Engholm DH, Kilian M, Goodsell DS, et al. A visual review of the human pathogen Streptococcus pneumoniae. FEMS Microbiol Rev 2017;41:854-79. [Crossref] [PubMed]
- Feldman C, Anderson R. The Role of Streptococcus pneumoniae in Community-Acquired Pneumonia. Semin Respir Crit Care Med 2020;41:455-69. [Crossref] [PubMed]
- Varghese R, Jayaraman R, Veeraraghavan B. Current challenges in the accurate identification of Streptococcus pneumoniae and its serogroups/serotypes in the vaccine era. J Microbiol Methods 2017;141:48-54. [Crossref] [PubMed]
- Ikken Y, Charof R, Elouennass M, et al. The novel biphasic medium for transport, culture and conservation at an ambient temperature of Neisseria meningitidis, Streptococcus pneumoniae and Haemophilus influenzae. World J Microbiol Biotechnol 2021;37:187. [Crossref] [PubMed]
- Webber DM, Wallace MA, Burnham CA, et al. Evaluation of the BioFire FilmArray Pneumonia Panel for Detection of Viral and Bacterial Pathogens in Lower Respiratory Tract Specimens in the Setting of a Tertiary Care Academic Medical Center. J Clin Microbiol 2020;58:e00343-20. [Crossref] [PubMed]
- Murdoch DR, O’Brien KL, Scott JA, et al. Breathing new life into pneumonia diagnostics. J Clin Microbiol 2009;47:3405-8. [Crossref] [PubMed]
- Murdoch DR. How recent advances in molecular tests could impact the diagnosis of pneumonia. Expert Rev Mol Diagn 2016;16:533-40. [Crossref] [PubMed]
- Poulain D. Candida albicans, plasticity and pathogenesis. Crit Rev Microbiol 2015;41:208-17. [Crossref] [PubMed]
- Murphy CN, Fowler R, Balada-Llasat JM, et al. Multicenter Evaluation of the BioFire FilmArray Pneumonia/Pneumonia Plus Panel for Detection and Quantification of Agents of Lower Respiratory Tract Infection. J Clin Microbiol 2020;58:e00128-20. [Crossref] [PubMed]
- Yugueros-Marcos J, Barraud O, Iannello A, et al. New molecular semi-quantification tool provides reliable microbiological evidence for pulmonary infection. Intensive Care Med 2018;44:2302-4. [Crossref] [PubMed]
- Monard C, Pehlivan J, Auger G, et al. Multicenter evaluation of a syndromic rapid multiplex PCR test for early adaptation of antimicrobial therapy in adult patients with pneumonia. Crit Care 2020;24:434. [Crossref] [PubMed]


