Prediction of obstructive sleep apnea using deep learning in 3D craniofacial reconstruction
Introduction
Obstructive sleep apnea (OSA) is a common sleep disorder characterized by the collapse of the upper airway during sleep, resulting in an increased risk of multisystem dysfunction and damage to the cardiovascular and endocrine systems. The prevalence of OSA is increasing annually, leading to a significant medical burden and economic cost (1). OSA affects 9% to 38% of the global population (2), with a prevalence exceeding 50% in some countries (3). In addition, some studies have shown that the prevalence of moderate to severe sleep-disordered breathing [apnea-hypopnea index (AHI) ≥15 events/h] is 23.4% in women and 49.7% in men over 40 years of age (4). Polysomnography (PSG) is currently the gold standard for diagnosing OSA (5). However, it requires patients to sleep through the night in a specialized sleep laboratory and a professional technician to manually determine OSA based on signals such as airflow, electroencephalogram (EEG), and eye movements, which are very time-consuming and costly (6). These limitations result in many patients with potential OSA remaining undiagnosed (7).
Various degrees of obesity as well as craniofacial and upper airway structural abnormalities, including maxillary deficiency and retro-position (8,9), the inferior position of the hyoid bone (10,11), a larger tongue, and soft palate volume (12,13), cause OSA. Upper airway computed tomography (CT) is frequently used to assess airway structural abnormalities in patients with OSA; however, this also requires manual measurements by trained personnel (14,15). A simple photographic method for craniofacial phenotyping has been developed (16). This method was demonstrated to have predictive utility in identifying patients with OSA (17). Facial photography can provide a composite measure of both skeletal and soft tissues and has been shown to capture phenotypic information regarding upper airway structures (18,19). Hence, the presence of OSA or even the magnitude of the AHI can be predicted using facial imaging with an accuracy that can reach 87–91% (20,21). However, for this technique, the faces of patients often need to be individually outlined, photographed, and measured.
Based on these methods and anatomical principles, we aimed to develop a method using artificial intelligence (AI) and upper airway CT to automatically predict OSA without manual measurement and test its accuracy. We aimed to assess the predictive utility of these new measurements as a powerful noninvasive tool for providing an early warning of OSA, even when the patient is undergoing a CT scan for other diseases. We present the following article in accordance with the STARD reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-734/rc).
Methods
Patients
For inclusion in this study, we retrospectively and randomly selected 219 patients with OSA (AHI ≥10/h) and 81 controls (AHI <10/h) aged 18 years or older from patients who visited Beijing Tongren Hospital between July 2017 and February 2021 and who subsequently underwent PSG and upper airway CT. Patients with central sleep syndrome, a history of facial deformity, upper airway surgery or facial surgery were excluded.
CT images were mainly retrospectively collected from patients who visited our hospital complaining of sleep apnea. They underwent PSG to confirm the diagnosis of OSA, indicated a desire to undergo surgical treatment, and underwent preoperative upper airway CT.
CT images of individuals in the control group were mainly derived from patients who came to our hospital with a diagnosis of leukoplakia of the vocal cords and were going to be treated surgically. These patients underwent preoperative CT of the neck to define the extent of the lesion. PSG was performed to clarify whether these patients had OSA so that the risk from the subsequent anesthesia could be assessed. The control group was mainly selected retrospectively from this group of patients with a PSG diagnosis and an AHI less than 10. If the AHI was greater than or equal to 10, the patient was included in the experimental group.
This clinical study was a retrospective study, which only collected patients’ clinical data; it did not interfere with patients’ treatment plans and did not pose any risk to patients’ physiology. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Beijing Tongren Hospital, Beijing, China (No. TRECKY2019-049). Informed consent was waived due to the retrospective nature of the study.
Anthropometric measurements
Clinical variables, including height (m), weight (kg), body mass index (BMI, weight/height2, kg/m2), and neck circumference (circumference at the level of the thyroid cartilage angle, cm) were collected.
PSG
All participants underwent standard overnight polysomnographic monitoring, which included EEGs (C3/A2 and C4/A1, measured using surface electrodes), electrooculograms (measured using surface electrodes), submental electromyograms (measured by surface electrodes), nasal airflow (measured using a nasal cannula with a pressure transducer), oral airflow (measured with a thermistor), chest wall and abdominal movements (recorded by inductance plethysmography), electrocardiography, and pulse oximetry. Respiratory events were classified according to the American Academy of Sleep Medicine Criteria 2012 (version 2.0) (22). Apnea was defined as a drop in peak signal excursion by ≥90% of the pre-event baseline over ≥10 s with an oronasal thermal sensor. Hypopnea was defined as a drop in peak signal excursions by ≥30% of the pre-event baseline over ≥10 s, using nasal pressure in association with either ≥3% arterial oxygen desaturation or arousal. AHI was calculated as the total duration of all apneic and hypopneic events divided by the total sleep time. We chose AHI ≥10 as an index for the diagnosis of OSA.
Upper airway CT analysis
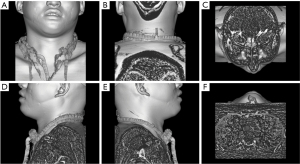
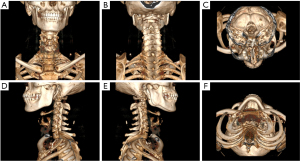
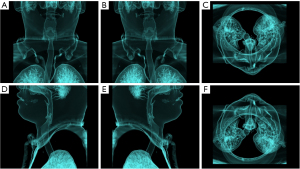
Upper airway CT scans were performed in the supine position at end-expiration using the Frankfort plane perpendicular to the horizontal plane while the patients were awake. Throughout the scans, the patients were instructed to keep their mouths closed without swallowing or chewing. Each patient had a complete CT sequence that included 300–600 CT images with one 512×512 pixel channel per CT image. We reconstructed each patient’s CT using Mimics image processing software version 21.0 (Materialise, Leuven, Belgium) with the following 3 methods: the skin method (Figure 1), the skeletal method (Figure 2), and the airway method (Figure 3) We intersected the reconstructed 3D model in 6 directional views (front, back, top, bottom, left profile, and right profile; Figures 1-3), using six 2D images to represent the information on a complete 3D model. Each image had 3 channels of 442×442 pixels.
Data preprocessing
Figure 4 illustrates the structure of the neural networks. After reconstruction with each of the 3 different methods, we used CT sequences to obtain 3D models and acquired 6 views in each direction (anterior, posterior, superior, inferior, left contour, and right contour). The 6 views from each reconstruction method and all 18 views were imported into the model to determine whether the patient experienced OSA. The task was converted into a binary classification of six 2D images. We chose ResNet-18 (22) as the back end of this model on the PyTorch framework. Each model was trained using the AdamW (23) optimizer with an initial learning rate of with a cosine learning rate decay schedule of . k is the current training step, and K is the total training step. The other hyperparameters are listed in Table S1. We chose the loss function as a cross-entropy function with weight. As shown in Eq. [1], we assigned a higher weight to the loss function for normal individuals, which was set to 2.
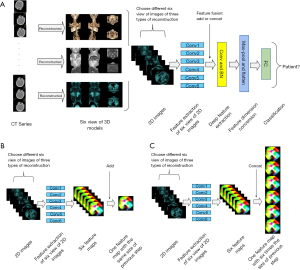
We set the first layer of the network to 6 different convolutional layers to simultaneously extract features from the 6 images (when importing all 18 views into the network, the first layer was set to 18 convolutional layers), allowing it to be applied to multiple input images. The features of the initially captured images were then fused using 2 types of feature fusion methods: Add and Concat. Add is a commonly used feature fusion method, as shown in Figure 4B. Each of the views was imported into a separate convolutional layer for initial feature extraction to obtain a feature map. The Add feature fusion method directly adds these feature maps to obtain a feature map of the same size as the original feature map and contains the feature information on the maps. Concat differs from Add in that it obtains each feature map of the same shape and directly stitches them together in a certain dimension to obtain a new feature map (Figure 4C).
The features fused by Concat or Add methods were imported into the convolutional layer for feature extraction. After several feature extractions, the feature map was pooled for the maximum value, and the pooled feature dimensions were converted into flat vectors for input to the fully connected layer. Finally, the fully connected layer output a 2D probability vector indicating whether or not the patient had OSA. We used 5-fold cross-validation for training, validation, and testing to reduce overfitting and to obtain as much valid information as possible. The method was as follows: we divided the dataset into 5 equal parts and then selected 1 of these parts as the test set and the rest as training and verification sets. We performed training and testing 5 times and finally reported the performance as the average of the 5 results.
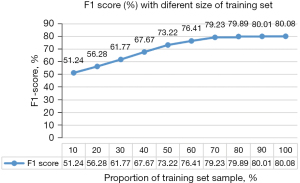
In addition, we selected different sample sizes for training to obtain F1 values and thus determine whether the sample size was sufficient. We randomly selected 20% of the data as the test set and 10% to 100% of the remaining data as the training set. The training mode included all 18 views plus the fusion method.
The model was trained using a Cor 4210 CPU (Intel, CA, USA) and GeForce GTX 3090 GPU (Nvidia, CA, USA).
Statistical analysis
To evaluate the performance of the model, the following parameters were calculated: area under the receiver operating characteristic (AUC) curve, precision, recall, and F1 score.
Results
There was a significant difference in BMI between the OSA and non-OSA patients, and no significant difference in age or male-to-female ratio was observed (Table S2).
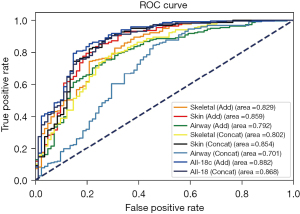
We experimented with each of the 6 views obtained through these 3 modes. For every type of reconstruction, we used 2 methods for feature fusion: Add and Concat. In addition, we imported all 18 views into a single model. The experimental results are shown in Table S3 and Figure 5. The best performance was achieved when using all 18 views with Add as the feature fusion method compared with the other reconstruction and fusion methods. Using all 18 views with the Add fusion approach provided the best performance for this prediction method, with the largest AUC of 0.882. This was followed by using all 18 views with the Concat fusion method, resulting in an AUC of 0.868. The skin reconstruction method was the best predictor of OSA using only 6 images and had an AUC of 0.859. Airway reconstruction using the Concat fusion method had the lowest AUC (0.701).
The results of the sample size calculation are shown in Figure 6. The model’s performance did not improve significantly when the training set exceeded 154 (70%) of the data, proving that our dataset’s size was sufficient. In addition, the times (seconds) for the prediction of OSA with the different methods are listed in Table S4.
Discussion
To our knowledge, this is the first study to use CT-based deep learning to predict OSA. This program is a convenient tool that can be used as a plug-in for various image-examination software programs. When a patient undergoes a head and neck CT examination, the program can automatically indicate whether the patient has OSA without requiring manual measurement or assessment. The clinician receives an alert and determines whether further diagnostic testing is required in the context of the actual situation. Our model is sufficiently simple for routine clinical use and shows good performance, which will help increase OSA screening rates and reduce screening costs.
Convolutional neural networks perform well in image classification tasks, and there are many common neural network models, such as ResNet (22), InceptionNet (24), and DeseNet (25), that perform well in image classification tasks. Compared with the other two neural network models, ResNet-18 has the advantages of a simple network structure and a wide range of applications with the residual structure, which makes it possible to avoid overfitting. We modified the structure based on the original ResNet-18 model by setting the first layer of the network to 6 (or 18) different convolutional layers to extract the features of 6 images in parallel so that it can be applied to multiple input images to reduce the parameters of the model and the training time. After the extraction, we performed further feature extraction by fusing the features of the initially extracted images using 2 different methods. In this manner, we could make good use of the information in the images without significantly increasing the model parameters and training time.
Our findings showed that using all 18 views was the most effective predictor of OSA with an accuracy rate of 88.2%, probably because this method contains the most information (including facial features, bones, upper airway morphology, etc.). The skin reconstruction method was the best predictor of OSA using only 6 images, with an accuracy of 85.9%. The skin may contain information related to age or obesity (e.g., skeleton size and subcutaneous fat thickness) (26,27). AI can identify and use this information; however, the naked eye often fails to do so.
Recently, many studies have applied machine learning (ML) methods to predict whether an individual has OSA. Pombo et al. (28) reported that ML accounted for 85.25% of the methods used to predict OSA, with good performance and reliability, and was the most used method and the most popular algorithm. The questionnaire is a convenient assessment tool that usually includes symptoms, demographic variables, and other factors. Clinicians frequently use it for the initial assessment of the probability of a patient having OSA and for deciding whether further diagnostic testing is required. Tan et al. (29) reported that the NoSAS (Neck circumference, Obesity, Snoring, Age, Sex) score was similar to the STOP-BANG (snoring history, tired during the day, observed stop breathing while sleep, high blood pressure, BMI more than 35 kg/m2, age more than 50 years, neck circumference more than 40 cm and male gender) and Berlin questionnaires in predicting OSA. Tan et al. (29) concluded that the NoSAS score was slightly better than that of other questionnaires. However, regardless of the effect, they all agreed that the questionnaire could be used as a screening tool. Eijsvogel et al. (30) modified the scoring system to predict OSA using a two-step method, with a sensitivity of 63.1% and a specificity of 90.1%. Rowley et al. (31) evaluated 4 previously published clinical prediction models based primarily on symptoms (including snoring, witnessed apneas, and gasping/choking) and anthropometric data (including BMI, age, and sex) for predicting OSA, reporting a sensitivity range of 33–39% and a specificity range of 87–93%. Overall, questionnaires and anthropometrics have low levels of accuracy and greater patient subjectivity, which limits their use in clinical models. Therefore, we do not advocate the use of clinical tools, questionnaires, or predictive algorithms to diagnose OSA in adults (32).
Many studies used a combination of multiple PSG signals (e.g., electrocardiogram, EEG, and SpO2) to predict OSA with high accuracy and good predictive power (33). Compared with questionnaires, these methods are based on objective measurements with fewer subjective biases. However, they still require complex monitoring and are unsuitable for OSA screening.
The advent of photographic analysis has provided a replacement technique for the quantitative assessment of craniofacial morphology and prediction of OSA. Reports show that the accuracy of OSA detection using photographic variables can reach 76.1% (16,17). However, strict standardization of calibration photography (control of distance from the camera and contour positions), manual annotation, and measurement may be both time-consuming and tedious, hindering the application of this technique (21). With the development of AI, deep learning has been highly successful in tasks such as the detection, segmentation, and classification of objects for both images and videos (34). OSA can be predicted simply and quickly using deep learning.
Combining facial features with AI provides objective measurement information and eliminates the need for manual measurements, making it a reliable method for the large-scale screening and early-warning provision of OSA. Tabatabaei et al. (35) separated the frontal and lateral perspectives of the face from the overall image and achieved an accuracy of up to 61.8% in their test set. Brink-Kjaer et al. (8) used frontal and lateral facial photographs and input them directly into a neural network classifier to achieve an average accuracy of 62% for OSA detection. Islam et al. (36) transformed 3D scans into frontal 2D depth maps. Three different ConvNet neural network models were chosen to classify patients, demonstrating 57.1–67.4% accuracy in the test group. Nosrati et al. (37) achieved 73.3% accuracy in predicting OSA using a support vector machine (SVM). Lee et al. (17) achieved 48.2% sensitivity and 92.4% specificity in predicting OSA using a single photographic measurement (mandibular width-length angle) and the simplest classification and regression trees (CART) model. They also applied CART using 4 photographic measurements (mandibular width-length angle, neck depth, mandible width, face width-lower face depth angle) to predict OSA with a sensitivity of 70.2% and a specificity of 87.9% (17). Similar sensitivities and specificities were obtained by Espinoza-Cuadros et al. (38) using Lee et al.’s facial features (specificity of 79.1% and sensitivity of 85.1%). He et al. (21) reported the best results thus far using frontal, 45-degree lateral, and 90-degree lateral images to predict OSA using ConvNet neural network models and could attain 91–95% sensitivity and 73–80% specificity. Hanif et al. (39) detected predefined facial landmarks and aligned the scans in 3D space. The scans were subsequently rendered and rotated in 45-degree increments to generate 2D images and depth maps that were fed into a convolutional neural network to predict the AHI values from the PSG. The mean absolute error of the proposed model was 11.38 events/hour, with an accuracy of 67% (39). The drawback of this prediction method is that it requires the facial acquisition of individual patients and needs to be calibrated and uploaded, which makes it difficult to collect patient data. Further, the upload of facial photographs requires consideration of data security and patient privacy protection.
Our model assesses objective data and does not include human subjectivity in the diagnostic process. It is also quite simple to apply in the clinic, does not require additional calibrations and acquisition of patients’ facial photographs, and is less labor-intensive and time-consuming. Moreover, this early detection allows for the OSA screening of a broader population than simply individuals who wish to be photographed. It minimizes the delay in diagnosis and referral of patients to secondary or tertiary care without the need to consider patient privacy breaches. Only Tsuiki et al. (20) reported a similar study in which they developed a deep convolutional neural network using lateral cephalometric radiographs to diagnose severe OSA, with sensitivity reaching 92%.
This study has some limitations. First, the patients were all recruited from a sleep center population, and this population may have a higher prevalence of OSA, males, and overweight individuals than does the general population. This is a limitation of the present and other similar studies. Second, our model requires further training in a female population due to the limited number of female patients and possible craniofacial differences between the sexes (40). In addition, our study included only Chinese individuals, and generalizability to other ethnicities is uncertain. Moreover, we only included patients with moderate and high OSA; however, previous studies have shown that an AHI threshold of 5 events/h is where concomitant changes in facial morphology are most remarkable (41). CT reconstruction includes clear identifiers (skin) and rich health data that require the same information protection as facial photography. In the future, further model refinement and validation with other ethnic groups and community populations are needed to overcome these limitations. Applying our model to populations that include mild OSA samples and more females will provide a more practical model for predicting OSA. In this study, the model did not include causative factors for OSA other than craniofacial anatomy (e.g., upper airway muscle responsiveness, respiratory control, arousal capacity). Further inclusion of physiological respiratory factors could be considered to improve the predictive power of the model; otherwise, false positives arising from low accuracy could lead to a more costly investigation. However, false positives in patients may also indicate the presence of potentially hazardous anatomic features that need to be followed closely. This is because it is still possible to have OSA as weight and age increase. Due to technical limitations, we used 2D photographs from different angles to represent the 3D structure of the face, but this may still not cover all facial shape and contour information. Using 3D facial scans for direct OSA prediction may be a future trend. In addition, 3D segmentation of the airway, combined with sleep apnea detection parameters, could be used in the diagnosis and prediction of OSA, which may be informative and lead to better results.
Conclusions
We propose a model for predicting OSA using upper airway CT and deep learning. The model has satisfactory performance and has high potential utility in enabling CT to accurately identify patients with moderate to severe OSA. It can be further incorporated into CT procedures to alert clinicians of OSA in patients who undergo head and neck CT. It can serve as an effective screening tool for OSA in clinics or communities to guide further treatment choices, contributing to epidemiological investigations and health management.
Acknowledgments
Funding: This research was supported by the Ministry of Industry and Information Technology of China (No. 2020-0103-3-1), National Natural Science Foundation of China (No. 81970866), and Beijing Municipal Administration of Hospitals’ Youth Programme (Code: *QMS20190202*).
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-734/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-734/dss
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-734/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-734/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Beijing Tongren Hospital, Beijing, China (No. TRECKY2019-049). Informed consent was waived due to the retrospective nature of the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lee W, Nagubadi S, Kryger MH, et al. Epidemiology of Obstructive Sleep Apnea: a Population-based Perspective. Expert Rev Respir Med 2008;2:349-64. [Crossref] [PubMed]
- Senaratna CV, Perret JL, Lodge CJ, et al. Prevalence of obstructive sleep apnea in the general population: A systematic review. Sleep Med Rev 2017;34:70-81. [Crossref] [PubMed]
- Benjafield AV, Ayas NT, Eastwood PR, et al. Estimation of the global prevalence and burden of obstructive sleep apnoea: a literature-based analysis. Lancet Respir Med 2019;7:687-98. [Crossref] [PubMed]
- Heinzer R, Vat S, Marques-Vidal P, et al. Prevalence of sleep-disordered breathing in the general population: the HypnoLaus study. Lancet Respir Med 2015;3:310-8. [Crossref] [PubMed]
- Kushida CA, Littner MR, Morgenthaler T, et al. Practice parameters for the indications for polysomnography and related procedures: an update for 2005. Sleep 2005;28:499-521. [Crossref] [PubMed]
- Ibáñez V, Silva J, Cauli O. A survey on sleep questionnaires and diaries. Sleep Med 2018;42:90-6. [Crossref] [PubMed]
- Young T, Peppard PE, Gottlieb DJ. Epidemiology of obstructive sleep apnea: a population health perspective. Am J Respir Crit Care Med 2002;165:1217-39. [Crossref] [PubMed]
- Brink-Kjaer A, Olesen AN, Peppard PE, et al. Automatic detection of cortical arousals in sleep and their contribution to daytime sleepiness. Clin Neurophysiol 2020;131:1187-203. [Crossref] [PubMed]
- Piriyajitakonkij M, Warin P, Lakhan P, et al. SleepPoseNet: Multi-View Learning for Sleep Postural Transition Recognition Using UWB. IEEE J Biomed Health Inform 2021;25:1305-14. [Crossref] [PubMed]
- Riha RL, Brander P, Vennelle M, et al. A cephalometric comparison of patients with the sleep apnea/hypopnea syndrome and their siblings. Sleep 2005;28:315-20.
- Hsu PP, Tan AK, Chan YH, et al. Clinical predictors in obstructive sleep apnoea patients with calibrated cephalometric analysis--a new approach. Clin Otolaryngol 2005;30:234-41. [Crossref] [PubMed]
- Lowe AA, Santamaria JD, Fleetham JA, et al. Facial morphology and obstructive sleep apnea. Am J Orthod Dentofacial Orthop 1986;90:484-91. [Crossref] [PubMed]
- Ferguson KA, Ono T, Lowe AA, et al. The relationship between obesity and craniofacial structure in obstructive sleep apnea. Chest 1995;108:375-81. [Crossref] [PubMed]
- Schwab RJ, Pasirstein M, Pierson R, et al. Identification of upper airway anatomic risk factors for obstructive sleep apnea with volumetric magnetic resonance imaging. Am J Respir Crit Care Med 2003;168:522-30. [Crossref] [PubMed]
- O'Toole L, Quincey VA. Obstructive sleep apnoea. Lancet 2002;360:2079. [Crossref] [PubMed]
- Lee RW, Chan AS, Grunstein RR, et al. Craniofacial phenotyping in obstructive sleep apnea--a novel quantitative photographic approach. Sleep 2009;32:37-45.
- Lee RW, Petocz P, Prvan T, et al. Prediction of obstructive sleep apnea with craniofacial photographic analysis. Sleep 2009;32:46-52.
- Lee RW, Sutherland K, Chan AS, et al. Relationship between surface facial dimensions and upper airway structures in obstructive sleep apnea. Sleep 2010;33:1249-54. [Crossref] [PubMed]
- Sutherland K, Schwab RJ, Maislin G, et al. Facial phenotyping by quantitative photography reflects craniofacial morphology measured on magnetic resonance imaging in Icelandic sleep apnea patients. Sleep 2014;37:959-68. [Crossref] [PubMed]
- Tsuiki S, Nagaoka T, Fukuda T, et al. Machine learning for image-based detection of patients with obstructive sleep apnea: an exploratory study. Sleep Breath 2021;25:2297-305. [Crossref] [PubMed]
- He S, Su H, Li Y, et al. Detecting obstructive sleep apnea by craniofacial image-based deep learning. Sleep Breath 2022;26:1885-95. [Crossref] [PubMed]
- He K, Zhang X, Ren S, et al. Deep Residual Learning for Image Recognition. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2016:770-8.
- Loshchilov I, Hutter F. Decoupled weight decay regularization. arXiv preprint 2017. doi:
10.48550 /arXiv.1711.05101. - Szegedy C, Liu W, Jia Y, et al. Going deeper with convolutions. Proceedings of the IEEE conference on computer vision and pattern recognition; 2015:1-9.
- Huang G, Liu Z, Pleiss G, et al. Convolutional Networks with Dense Connectivity. IEEE Trans Pattern Anal Mach Intell 2022;44:8704-16. [Crossref] [PubMed]
- Kamei T, Aoyagi K, Matsumoto T, et al. Age-related bone loss: relationship between age and regional bone mineral density. Tohoku J Exp Med 1999;187:141-7. [Crossref] [PubMed]
- Satoh M, Mori S, Nojiri H, et al. Age-associated changes in the amount of subcutaneous tissue in the face evaluated in the ultrasonic B mode. J Soc Cosmet Chem Jpn 2004;38:292-8.
- Pombo N, Garcia N, Bousson K. Classification techniques on computerized systems to predict and/or to detect Apnea: A systematic review. Comput Methods Programs Biomed 2017;140:265-74. [Crossref] [PubMed]
- Tan A, Hong Y, Tan LWL, et al. Validation of NoSAS score for screening of sleep-disordered breathing in a multiethnic Asian population. Sleep Breath 2017;21:1033-8. [Crossref] [PubMed]
- Eijsvogel MM, Wiegersma S, Randerath W, et al. Obstructive Sleep Apnea Syndrome in Company Workers: Development of a Two-Step Screening Strategy with a New Questionnaire. J Clin Sleep Med 2016;12:555-64. [Crossref] [PubMed]
- Rowley JA, Aboussouan LS, Badr MS. The use of clinical prediction formulas in the evaluation of obstructive sleep apnea. Sleep 2000;23:929-38. [Crossref] [PubMed]
- Ramar K, Dort LC, Katz SG, et al. Clinical Practice Guideline for the Treatment of Obstructive Sleep Apnea and Snoring with Oral Appliance Therapy: An Update for 2015. J Clin Sleep Med 2015;11:773-827. [Crossref] [PubMed]
- Roebuck A, Monasterio V, Gederi E, et al. A review of signals used in sleep analysis. Physiol Meas 2014;35:R1-57. [Crossref] [PubMed]
- Esteva A, Robicquet A, Ramsundar B, et al. A guide to deep learning in healthcare. Nat Med 2019;25:24-9. [Crossref] [PubMed]
- Tabatabaei Balaei A, Sutherland K, Cistulli P, et al. Prediction of obstructive sleep apnea using facial landmarks. Physiol Meas 2018;39:094004. [Crossref] [PubMed]
- Islam SMS, Mahmood H, Al-Jumaily AA, et al. Deep learning of facial depth maps for obstructive sleep apnea prediction. International Conference on Machine Learning and Data Engineering (iCMLDE) 2018:154-7.
- Nosrati H, Sadr N, Chazal PD. Apnoea-Hypopnoea Index Estimation using Craniofacial Photographic Measurements. Computing in Cardiology Conference IEEE 2016;43:297-381.
- Espinoza-Cuadros F, Fernández-Pozo R, Toledano DT, et al. Speech Signal and Facial Image Processing for Obstructive Sleep Apnea Assessment. Comput Math Methods Med 2015;2015:489761. [Crossref] [PubMed]
- Hanif U, Leary E, Schneider L, et al. Estimation of Apnea-Hypopnea Index Using Deep Learning On 3-D Craniofacial Scans. IEEE J Biomed Health Inform 2021;25:4185-94. [Crossref] [PubMed]
- Popovic RM, White DP. Influence of gender on waking genioglossal electromyogram and upper airway resistance. Am J Respir Crit Care Med 1995;152:725-31. [Crossref] [PubMed]
- Eastwood P, Gilani SZ, McArdle N, et al. Predicting sleep apnea from three-dimensional face photography. J Clin Sleep Med 2020;16:493-502. [Crossref] [PubMed]