
In lung adenocarcinoma, low expression of the cell surface extracellular nucleotidase CD39 is related to immune infiltration and a poor prognosis
Highlight box
Key findings
• The increased expression of CD39 can significantly prolong the prognosis of patients with lung adenocarcinoma. At the same time, immune escape may occur by inhibiting tumor cells in the tumor microenvironment of lung adenocarcinoma.
What is known and what is new?
• CD39 is a key molecule in the adenosine pathway.
• Current research shows that CD39 is also involved in different biological events of various tumors, such as AML, glioma and other tumors. At the same time, the prediction of the mechanism is added, which provides a reference for later research.
What is the implication, and what should change now?
• Basic experiments in vivo and in vitro are added to clarify the mechanism of CD39 in lung adenocarcinoma.
Introduction
Lung cancer has one of the highest proportions of occurrence and mortality among human cancers. Lung adenocarcinoma constitutes 30–35% of all lung cancer cases and is a type of non-small cell lung cancer (NSCLC), which is the prevalent type of lung cancer (1). Patients with lung cancer have increased recently due to smoking and air pollution (2). According to studies, less than 15% of people with severe lung adenocarcinoma will survive for at least 5 years. The long-term survival rate for lung adenocarcinoma is still quite low despite the development of numerous new treatments, including targeted therapy and immunotherapy (3,4). Most patients are diagnosed at an advanced stage, which is one of the primary causes of poor survival. Early diagnosis is one of the best ways to increase a lung cancer patient’s chances of survival and prognosis (5-7). Thus, to develop novel treatment drugs to improve patient survival, it is crucial to comprehend the molecular mechanisms underlying the occurrence and development of lung adenocarcinoma and establish biomarkers that may be detected at the early stages of the disease.
Adenosine triphosphate (ATP) is the main form of energy storage and energy supply in cellular energy metabolism. Under physiological conditions, ATP mainly exists in cells, its concentration is 1–10 mmol/L, and its extracellular concentration is extremely low (10–100 nmol/L). Extracellular ATP (eATP) is a crucial extracellular signaling molecule that, in addition to performing metabolic functions, binds to purinergic P2 receptors as danger-associated molecular patterns (DAMPs) and triggers inflammation via chemotaxis (8). It binds to purinergic P2 receptor, activates inflammatory bodies and platelets through chemotaxis, participates in inflammation, and triggers thrombosis (9). As an immunosuppressant on the A2 and A3 receptors on the surface of immune cells, ATP can be digested into adenosine by cascade enzymes in the extracellular space, preventing the harmful action of ATP (10).
A vital component of the adenosine pathway is the cell surface enzyme extracellular nucleotidase CD39, commonly referred to as extracellular nucleoside triphosphate diphosphate hydrolase 1 (ENTPD1) (11). ATP is composed of a macromolecule called adenosine and three simpler phosphate radicals. The last two phosphate radicals have "high energy bonds" on which a large amount of chemical energy is stored. Therefore, compounds like ATP are also called high energy phosphides. When ATP terminal monophosphate bond breaks, it releases energy to enable cells to do work or complete other physiological functions. When the body needs, adenosine triphosphate is decomposed into adenosine diphosphate (ADP) and energy is released for the body to use. Although adenosine triphosphate is the only material directly supplying energy in the body, its content in the tissues is very small. When it is continuously consumed, it must be continuously decomposed by other energy substances to release energy for other re synthesis. This paragraph should read: it specifically hydrolyzes extracellular ATP to ADP and/or AMP. AMP is hydrolyzed to adenosine via CD73 (another key extracellular nucleotidase of adenosine pathway) and extracellular 5'- nucleotidase to play an immunomodulatory role (12). Recent research has demonstrated that CD39 is also involved in various biological processes in many cancers. For example, blocking TIGIT (T cell Ig and ITIM domain), CD39, and A2AR can increase natural killer (NK)-92 cell-mediated cytotoxicity in acute myeloid leukemia (AML) (13). Immune responses against tumors can be modulated by cells with increased expression (14). To avoid immunological responses, glioma stem cells elevate CD39 expression through SOX2 regulation (15). Overexpression of PD-1 and CD39 in tumor infiltrating lymphocytes compared with peripheral blood lymphocytes in triple negative breast cancer (16). Genetically driven CD39 expression can shape the function of CD8+ T cells in human tumor invasion (17). CD39 expression defines cell failure in CD8+ T cells infiltrating tumors, and a subset of tumor infiltrating CD8+ T cells marked by high expression of immunosuppressive ATP exonucleotidase CD39 (18). However, a detailed investigation into the expression, prognosis, and mechanism of CD39 in lung cancer is limited. Moreover, the role of CD39 in tumor immune infiltration in lung adenocarcinoma is unknown. The ability to quickly develop high-throughput sequencing and microarray technology has made it possible to screen successfully for lung adenocarcinoma gene expression variations, which is an effective and promising tool for screening diagnostic biomarkers of benign or malignant conditions (19,20). Through The Cancer Genome Atlas (TCGA) database, our study primarily examined CD39 expression and its association with immune cell infiltration, immune cell biomarkers, or non-adenocarcinoma immunological checkpoints to calculate the survival of patients with lung adenocarcinoma. We also carried out bioinformatics analysis and confirmation to explore the possible mechanism of action of CD39. We present the following article in accordance with the TRIPOD reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1696/rc).
Methods
Data analysis using TCGA and TIMER databases
We obtained messenger RNA (mRNA) expression data for distinct cancer types from the TCGA and Tumor IMmune Estimation Resource (TIMER) databases. The data was adjusted, and we then used the R program (The R Foundation for Statistical Computing, Vienna, Austria) for differential expression analysis of CD39. A P value of <0.05 was considered statistically significant.
Clinical characteristics of individuals with lung adenocarcinoma in TCGA database and the association between CD39
From TCGA database, we retrieved the clinical features of lung cancer patients, and we then compared and examined the relationship between CD39 and these clinical characteristics and prognosis. The expression of CD39 in lung cancer was also evaluated using the log-rank test and the Mantel-Cox test. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
The nomogram construction and assessment
We created nomograms based on the multivariate analysis findings to tailor the expected survival probability at 1, 3, and 5 years. The RMS R program was used to create nomograms comprising clinical characteristics connected to CD39 and calibration plots. In addition, we compared the predictive accuracy of nomograms and individual prognostic factors using the C-index.
The correlation between CD39 and immune cells in TCGA and TIMER databases
Through the TIMER and TCGA online searches, we thoroughly examined the expression of tumor-infiltrating immune cells. Additionally, we examined the correlation between CD39 expression levels, immune checkpoint expression levels, and immune cell infiltration in lung adenocarcinoma.
Gene set enrichment analysis (GSEA)
For our online functional analysis, we used Metascape (https://metascape.org/gp/index.html#/main/step1). Metascape now includes differential genes for quantitative measurements.
Statistical analysis
The web resources were used to automatically compute the statistical analysis for this study. A P value or log-rank P value of <0.05 was regarded as statistically significant.
Results
Decreased CD39 expression in lung adenocarcinoma
Our examination of the TCGA and the TIMER databases revealed that CD39 expression in tumor tissues varied (Figure 1A,1B). Further investigation revealed that lung adenocarcinoma tumor samples had decreased CD39 expression (Figure 1C). When the receiver operating characteristic (ROC) curve was constructed, the area under the curve (AUC) was determined to be 0.679, indicating that CD39 had a stronger predictive impact (Figure 1D).
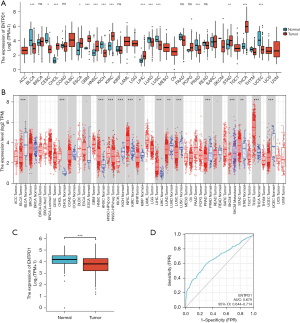
The prognosis of CD39 in lung adenocarcinoma
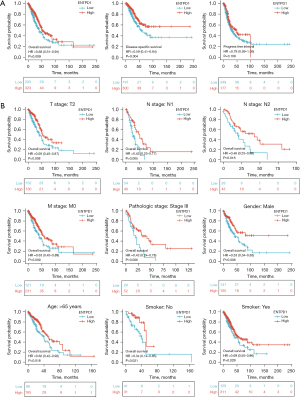
We observed that patients with high CD39 expression had better progression-free survival (PFS), disease-specific survival (DSS), and overall survival (OS) than those with low CD39 expression through TCGA database analysis (Figure 2A). Further stratified analysis showed that in T2, N1, N2, M0, pathological stage III, male gender, age >65 years, and smoking history of lung adenocarcinoma patients who also had high CD39 expression had a better prognosis (Figure 2B).
The association between CD39 in TCGA database and lung adenocarcinoma patient clinical information
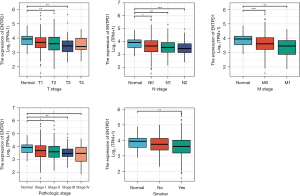
By examining the patient records in the TCGA database for lung adenocarcinoma, we determined that the expression of CD39 was variable in lung adenocarcinoma patients with distinct pathological stages, tumor-node-metastasis (TNM) stages, and smoking, whereas the expression of CD39 was low in tumor tissue. In comparison to normal tissues, lung adenocarcinoma patients with HCC who had low CD39 expression had worse survival in these different groups (Figure 3).
Nomogram construction
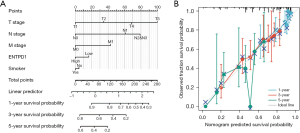
We developed nomograms based on the findings of multivariate analysis to determine 1-, 3-, and 5-year survival possibilities in lung adenocarcinoma patients, with a nomogram C-index of 0.688 (0.665–0.712) (Figure 4A). In the calibration plot, the bias correction line is near to the ideal curve (the 45-degree line), showing some consistency between projected and actual values (Figure 4B). As a result, this also demonstrated that the prediction model possesses a certain level of predictive performance.
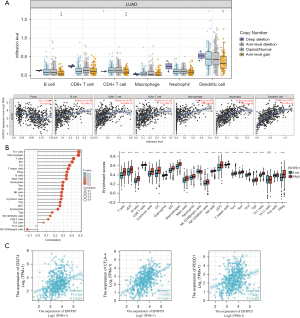
Immune cells and CD39 correlation in the TIMER and TCGA databases
We discovered that CD39 has a specific function in the immune system, and correlation analysis can offer important hints for researching the role and mechanism of CD39. As a result, we examined the relationship between CD39 and immune cell infiltration in lung adenocarcinomas using the TIMER database. CD39 expression was positively associated with B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells (DCs) (Figure 5A). Then, using TCGA database, we carried out pertinent analysis and verification. Correspondingly, we observed that CD39 was inversely connected to levels of infiltration of Th17 cells and NK CD56 bright cells but favorably correlated with levels of infiltration of Th1 cells, neutrophils, T cells, immature dendritic cell (iDCs), DCs, B cells, Treg, and T helper cells (Figure 5B). Subsequent investigation revealed that the immune checkpoint-related molecules PDCD1, CTLA-4, and CD274 had a positive correlation with CD39 expression. The differences were statistically significant (Figure 5C).
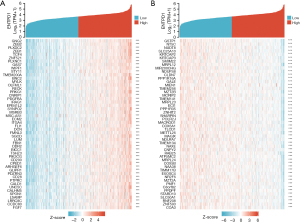
Screening of co-expressed genes of CD39
Genes associated with CD39 co-expression either favorably or negatively were found using data mining from TCGA platform. A graph displaying the top 50 lung cancer genes positively and negatively correlated with CD39 is provided in Figure 6A and Figure 6B.
Relationship between the expression of CD39 and other immunological markers
To further comprehend the connection between CD39 and immune responses, the TIMER dataset was used to examine the relationship between CD39 expression and many immunological markers in lung adenocarcinomas. Immune cells including DCs, NK cells, neutrophils, M2 macrophages, M1 macrophages, tumor-associated macrophages (TAM), monocytes, CD8+ T cells, T cells, and B cells were all characterized using the genes included in Table 1. In clinical cancer biopsies, tumor purity significantly impacts the architecture of immune infiltration. After correcting for tumor purity, lung adenocarcinoma immune cells’ expression of CD39 exhibited a favorable correlation with immunological markers.
Table 1
| Description | Gene markers | LUAD | ||||
|---|---|---|---|---|---|---|
| None | Purity | |||||
| Cor | P value | Cor | P value | |||
| B cell | CD19 | 0.416 | *** | 0.28 | *** | |
| CD79A | 0.458 | 0 | 0.342 | *** | ||
| T cell (general) | CD3D | 0.466 | *** | 0.324 | *** | |
| CD3E | 0.51 | *** | 0.378 | *** | ||
| CD2 | 0.532 | CD2 | 0.4111 | *** | ||
| CD8+ T cell | CD8A | 0.41 | 0 | 0.277 | *** | |
| CD8B | 0.349 | *** | 0.236 | *** | ||
| Monocyte | CD86 | 0.67 | 0 | 0.606 | *** | |
| CSF1R | 0.646 | *** | 0.585 | *** | ||
| TAM | CCL2 | 0.469 | 0 | 0.391 | *** | |
| CD68 | 0.503 | 0 | 0.431 | *** | ||
| IL-10 | 0.558 | *** | 0.479 | *** | ||
| M1 | IRF5 | 0.331 | *** | 0.242 | *** | |
| PTGS2 | 0.138 | *** | 0.148 | *** | ||
| NOS2 | 0.201 | *** | 0.131 | *** | ||
| M2 | CD163 | 0.55 | 0 | 0.482 | *** | |
| VSIG4 | 0.488 | 0 | 0.429 | *** | ||
| MS4A4A | 0.597 | 0 | 0.536 | *** | ||
| Neutrophils | CEACAM8 | 0.185 | *** | 0.183 | *** | |
| ITGAM | 0.533 | 0 | 0.469 | *** | ||
| CCR7 | 0.487 | 0 | 0.355 | *** | ||
| Dendritic cell | HLA-DPB1 | 0.474 | *** | 0.377 | *** | |
| Natural killer cell | KIR2DL1 | 0.061 | *** | 0.013 | *** | |
***, P<0.001. ENTPD1, extracellular nucleoside triphosphate diphosphate hydrolase 1 (ENTPD1 is the CD39); TIMER, Tumor IMmune Estimation Resource; LUAD, lung adenocarcinoma; Cor, correlation coefficient; TAM, tumor associated macrophages.
Additionally, we examined the connection between CD39 expression and different types of functional T cells, such as Th1, Th1-like, and Th2. After adjustment for tumor purity, we discovered that CD39 expression levels were positively linked with each of the 12 T-cell markers in lung adenocarcinoma (see Table 2 below).
Table 2
| Description | Gene markers | LUAD | ||||
|---|---|---|---|---|---|---|
| None | Purity | |||||
| Cor | P value | Cor | P value | |||
| Th1 | TBX21 | 0.369 | 0 | 0.235 | *** | |
| STAT4 | 0.419 | *** | 0.299 | *** | ||
| STAT1 | 0.397 | 0 | 0.31 | *** | ||
| TNF | 0.394 | 0 | 0.291 | *** | ||
| IFNG | 0.279 | *** | 0.161 | *** | ||
| Th1-like | CXCR3 | 0.389 | 0 | 0.262 | *** | |
| BHLHE40 | 0.168 | *** | 0.142 | *** | ||
| CD4 | 0.712 | *** | 0.652 | *** | ||
| Th2 | STAT6 | 0.052 | *** | 0.075 | *** | |
| STAT5A | 0.547 | *** | 0.462 | *** | ||
| Treg | FOXP3 | 0.571 | 0 | 0.474 | *** | |
| Resting Treg | IL2RA | 0.635 | *** | 0.57 | *** | |
***, P<0.001. ENTPD1, extracellular nucleoside triphosphate diphosphate hydrolase 1 (ENTPD1 is the CD39); TIMER, Tumor IMmune Estimation Resource; LUAD, lung adenocarcinoma; Cor, correlation coefficient.
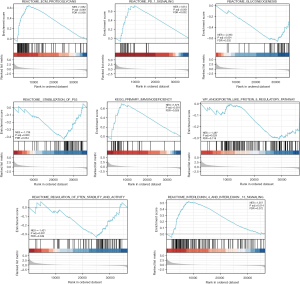
GSEA-based signaling pathways prediction
Using Metascape, we conducted a Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis online. The results revealed that CD39 might be engaged in
REACTOME_ECM_PROTEOGLYCANS,
KEGG_PRIMARY_IMMUNODEFICIENCY,
REACTOME_PD_1_SIGNALING,
REACTOME_GLUCONEOGENESIS,
REACTOME_STABILIZATION_OF_P53, REACTOME_REGULATION_OF_PTEN_STABILITY_AND_ACTIVITY,
REACTOME_INTERLEUKIN_4_AND_INTERLEUKIN_13_SIGNALING, and WP_ANGIOPOIETIN_LIKE_PROTEIN_8_REGULATORY_PATHWAY
to influence the biological events of lung adenocarcinoma, leading to different prognoses of lung adenocarcinoma (Figure 7).
Discussion
Since lung adenocarcinoma is one of the most prevalent cancer types and one of the malignant tumors with the maximum mortality and morbidity, understanding the molecular mechanism of CD39 may assist in the discovery of clinically effective therapies or exploration of compelling predictive biomarkers (21). Many studies have shown that CD39 is crucial to the development and progression of many cancers and diseases (22,23). Nevertheless, studies on CD39 in lung adenocarcinoma are relatively limited, and more investigation and exploration are required.
Through bioinformatics analysis, we investigated the significance of CD39 in the formation and progression of lung adenocarcinoma and the potential mechanism by which it interacts. We determined that CD39 expression was lowered in lung adenocarcinoma tumor tissue in the TCGA and TIMER databases and that lung adenocarcinoma patients with elevated CD39 expression had superior OS, DSS, and progression-free interval (PFI). In order to provide the prediction effectiveness and broader reference value for clinical application, we also developed a nomogram by evaluating the association between CD39 and clinical parameters of lung cancer patients.
Immune checkpoint therapies using anti-programmed death 1 (PD-1) and anti-programmed death ligand-1 (PD-L1) have transformed the management of lung adenocarcinoma and other metastatic malignancies (24). Even though immune checkpoint inhibitors can greatly improve the prognosis for lung adenocarcinoma patients, a substantial proportion of patients are resistant to them or they prove ineffective. Adenocarcinoma of the lung is considered one of the most immune tumors (25). In patients with lung adenocarcinoma, signals in the immunological microenvironment, such as the accumulation of metabolic byproducts or T cell malfunction, may greatly influence the response to immunosuppressive drugs. Prior research has demonstrated the widespread expression of CD39 in various disease-related immune cell types. However, the precise function of CD39 in these cells has been slowly revealed, such as how genetically determined CD39 expression can impact the activity of human tumor-infiltrating CD8+ T cells (17).
Poor survival is associated with interleukin 6 (IL-6)-induced CD39 expression on tumor-infiltrating NK cells in esophageal squamous cell carcinoma (26). AHR and CD39 cell expression in glioblastoma can control TAMs and T cells (27). In this study, we found that the expression of CD39 was positively correlated with immune invasion through the data analysis results in TIMER database and TCGA database, as well as the correlation with immune checkpoint related molecules PDCD1, CD274 and CTLA-4, and suggested that CD39 may play an important role in inhibiting tumor cells from immune escape in the lung adenocarcinoma tumor microenvironment. In this study, we discovered that the expression of CD39 was significantly associated with immune infiltration by evaluating the data from the TIMER and TCGA datasets. Moreover, the association with immune checkpoint-related markers PDCD1, CD274, and CTLA-4 demonstrated that CD39 plays a crucial function in the lung. The microenvironment of an adenocarcinoma tumor may have an important suppressive impact on the immune evasion of tumor cells. The findings of our further investigation into the relationship between CD39 and different immune markers revealed that CD39 was highly associated with all genes specifying immune cells and with different functional T cells, including Treg, Th1-like, and Th1, Resting Treg (regulatory T cells) characterization genes. Using GSEA as a foundation, we then inferred signaling pathways. CD39 may be involved in the extracellular matrix, immune microenvironment, PD-1 expression, glucose metabolism, PTEN stability, inflammation, and angiogenesis, among various other biological events in lung adenocarcinoma.
We assume that CD39 may be implicated in regulating multiple immune cells to establish an immunosuppressive tumor microenvironment based on these findings and the unique role of CD39 in the adenosine pathway. Although more in-depth research into the molecular processes by which CD39 prevents the development of an immunosuppressive environment should be conducted in both in vitro and in vivo conditions.
Conclusions
In summary, this work offers a wide range of data supporting the significance of CD39 in the genesis of lung adenocarcinoma and its perspective as a biomarker of the development of lung adenocarcinoma. By preventing tumor cells’ immunological escape from the tumor microenvironment, CD39 may enhance the prognosis for people with lung adenocarcinoma. Our results contribute to a potential target for lung adenocarcinoma anticancer treatments.
Acknowledgments
Funding: This study was supported by the Wu Jieping Foundation (No. 320.6750.19092-26).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1696/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1696/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wang J, He Z, Duan R. Expression of ASPM in Lung Adenocarcinoma and Its Relationship with Development and Prognosis. Zhongguo Fei Ai Za Zhi 2020;23:29-35. [Crossref] [PubMed]
- Zheng M. Classification and Pathology of Lung Cancer. Surg Oncol Clin N Am 2016;25:447-68. [Crossref] [PubMed]
- Kim CG, Kim KH, Pyo KH, et al. Hyperprogressive disease during PD-1/PD-L1 blockade in patients with non-small-cell lung cancer. Ann Oncol 2019;30:1104-13. [Crossref] [PubMed]
- Park CK, Cho HJ, Choi YD, et al. A Phase II Trial of Osimertinib in the Second-Line Treatment of Non-small Cell Lung Cancer with the EGFR T790M Mutation, Detected from Circulating Tumor DNA: LiquidLung-O-Cohort 2. Cancer Res Treat 2019;51:777-87. [Crossref] [PubMed]
- Yu Y, He J. Molecular classification of non-small-cell lung cancer: diagnosis, individualized treatment, and prognosis. Front Med 2013;7:157-71. [Crossref] [PubMed]
- Zengin T, Önal-Süzek T. Analysis of genomic and transcriptomic variations as prognostic signature for lung adenocarcinoma. BMC Bioinformatics 2020;21:368. [Crossref] [PubMed]
- Li M, Zhang C, Zhou L, et al. Identification and validation of novel DNA methylation markers for early diagnosis of lung adenocarcinoma. Mol Oncol 2020;14:2744-58. [Crossref] [PubMed]
- Trautmann A. Extracellular ATP in the immune system: more than just a "danger signal". Sci Signal 2009;2:pe6. [Crossref] [PubMed]
- Jacob F, Pérez Novo C, Bachert C, et al. Purinergic signaling in inflammatory cells: P2 receptor expression, functional effects, and modulation of inflammatory responses. Purinergic Signal 2013;9:285-306. [Crossref] [PubMed]
- Gessi S, Varani K, Merighi S, et al. Adenosine and lymphocyte regulation. Purinergic Signal 2007;3:109-16. [Crossref] [PubMed]
- Vijayan D, Young A, Teng MWL, et al. Targeting immunosuppressive adenosine in cancer. Nat Rev Cancer 2017;17:709-24. [Crossref] [PubMed]
- Hatfield SM, Kjaergaard J, Lukashev D, et al. Immunological mechanisms of the antitumor effects of supplemental oxygenation. Sci Transl Med 2015;7:277ra30. [Crossref] [PubMed]
- Brauneck F, Seubert E, Wellbrock J, et al. Combined Blockade of TIGIT and CD39 or A2AR Enhances NK-92 Cell-Mediated Cytotoxicity in AML. Int J Mol Sci 2021;22:12919. [Crossref] [PubMed]
- Shevchenko I, Mathes A, Groth C, et al. Enhanced expression of CD39 and CD73 on T cells in the regulation of anti-tumor immune responses. Oncoimmunology 2020;9:1744946. [Crossref] [PubMed]
- Liu B, Cao Y, Li Y, et al. Glioma Stem Cells Upregulate CD39 Expression to Escape Immune Response through SOX2 Modulation. Cancers (Basel) 2022;14:783. [Crossref] [PubMed]
- Zahran AM, Rayan A, Zahran ZAM, et al. Overexpression of PD-1 and CD39 in tumor-infiltrating lymphocytes compared with peripheral blood lymphocytes in triple-negative breast cancer. PLoS One 2022;17:e0262650. [Crossref] [PubMed]
- Gallerano D, Ciminati S, Grimaldi A, et al. Genetically driven CD39 expression shapes human tumor-infiltrating CD8(+) T-cell functions. Int J Cancer 2020;147:2597-610. [Crossref] [PubMed]
- Canale FP, Ramello MC, Núñez N, et al. CD39 Expression Defines Cell Exhaustion in Tumor-Infiltrating CD8(+) T Cells. Cancer Res 2018;78:115-28. [Crossref] [PubMed]
- Ding Y, Liu JH. The signature lncRNAs associated with the lung adenocarcinoma patients prognosis. Math Biosci Eng 2019;17:1593-603. [Crossref] [PubMed]
- Ran Z, Liu J, Wang F, et al. Analysis of Pulmonary Microbial Diversity in Patients with Advanced Lung Cancer Based on High-throughput Sequencing Technology. Zhongguo Fei Ai Za Zhi 2020;23:1031-8. [Crossref] [PubMed]
- You J, Yang G, Wu Y, et al. Plasma tRF-1:29-Pro-AGG-1-M6 and tRF-55:76-Tyr-GTA-1-M2 as novel diagnostic biomarkers for lung adenocarcinoma. Front Oncol 2022;12:991451. [Crossref] [PubMed]
- Li XY, Moesta AK, Xiao C, et al. Targeting CD39 in Cancer Reveals an Extracellular ATP- and Inflammasome-Driven Tumor Immunity. Cancer Discov 2019;9:1754-73. [Crossref] [PubMed]
- Laumont CM, Wouters MCA, Smazynski J, et al. Single-cell Profiles and Prognostic Impact of Tumor-Infiltrating Lymphocytes Coexpressing CD39, CD103, and PD-1 in Ovarian Cancer. Clin Cancer Res 2021;27:4089-100. [Crossref] [PubMed]
- Li CL, Hsia TC, Yang ST, et al. Efficacy of Prophylactic Traditional Chinese Medicine on Skin Toxicity of Afatinib in EGFR Mutation-Positive Advanced Lung Adenocarcinoma: A Single-Center, Prospective, Double-Blinded, Randomized-Controlled Pilot Trial. Integr Cancer Ther 2022;21:15347354221086663. [Crossref] [PubMed]
- Yan M, Yin X, Zhang L, et al. High expression of HOXB3 predicts poor prognosis and correlates with tumor immunity in lung adenocarcinoma. Mol Biol Rep 2022;49:2607-18. [Crossref] [PubMed]
- Zheng Y, Li Y, Tang B, et al. IL-6-induced CD39 expression on tumor-infiltrating NK cells predicts poor prognosis in esophageal squamous cell carcinoma. Cancer Immunol Immunother 2020;69:2371-80. [Crossref] [PubMed]
- Takenaka MC, Gabriely G, Rothhammer V, et al. Control of tumor-associated macrophages and T cells in glioblastoma via AHR and CD39. Nat Neurosci 2019;22:729-40. [Crossref] [PubMed]
(English Language Editor: J. Jones)


