
Correlation between oncogene integrator complex subunit 7 and a poor prognosis in lung adenocarcinoma
Highlight box
Key findings
• It has been proposed that INTS7 is an appropriate target for therapy and a prognostic marker for lung cancer.
What is known and what is new?
• There have been several INT subunits implicated in human cancer, the prognostic value of INTS7 in lung adenocarcinoma has been demonstrated in a recent study, linked INTS7 to tumor microenvironment and immunotherapy response.
• We know little about the relationship between INTS7 and lung adenocarcinoma in single cell resolution, as well as the underlying mechanisms.
What is the implication, and what should change now?
• There is evidence that INTS7 stimulates migration and invasion, promotes proliferation, and inhibits apoptosis in lung adenocarcinoma cells, however the further mechanism and signaling pathway should be explored in deeply.
Introduction
Lung adenocarcinoma (LUAD) is one of the most prevalent and malignant tumors worldwide, and it continues to have the lowest 5-year survival rate among the major cancers (1-3). Lung cancer can be divided into the following 2 types: (I) small cell lung cancer (SCLC); and (II) non-small cell lung cancer (NSCLC). The 2 types differ in their genetic causes and responses to treatment in clinical settings. SCLC often responds well to chemotherapy at the start of treatment, while NSCLC is less sensitive to chemotherapy (4). LUAD is a subtype of NSCLC, which accounts for approximately 50% of lung cancers and has had an increased incidence in recent decades (5). Based on a better understanding of tumor biology, the identification and use of biomarkers for the management of NSCLC patients have become a research trend. An increasing amount of effort is being made to identify the subtypes of NSCLC that might respond well to specific therapies. However, because of acquired resistance, the long-term benefits of immunotherapies driven by various tumor biomarkers are severely limited (6).
Immunotherapy and targeted therapy have been used to treat LUAD; however, the 5-year survival rate of LUAD patients remains unsatisfactory (7). Currently, CEA (carcinoembryonic antigen) is the main LUAD tumor marker used in clinical settings. However, due to its lack of specificity and sensitivity, it cannot be used to make reliable evaluations in diagnosing LUAD (8). Thus, potential therapeutic targets and prognostic biomarkers for LUAD patients need to be identified (9,10).
The integrator complex (INT) is one of the major components of ribonucleic acid (RNA) polymerase II-mediated transcription machinery, and is involved in regulating most dependent genes. INT forms by assembling at least 14 distinct subunits and based on their molecular weights, each subunit has been named INTS1-INTS14 (11). Studies have revealed that some INT subunits may also be involved in human cancer (12). A small subset of INT genes was found to be deregulated in several cancer types; for example, the integrator complex subunit 7 (INTS7) is significantly overexpressed in several cancers (11). INTS7 mediates the processing of small-nuclear RNA (snRNA) and is associated with RNA polymerase II (13). Many researchers have reported alternatively spliced transcript variants of INTS7 encoding multiple isoforms (14). INST7 has been shown to be highly mutated and significantly overexpressed in various human cancers; for example, INST7 expression has been shown to be increased in breast cancer, cholangiocarcinoma, colon cancer, and liver and lung squamous cell carcinoma, and the INTS7 mutation has been shown to be associated with endometrial cancer (11).
The advent of single-cell–RNA sequencing technologies has enabled the comprehensive analysis of individual cells in organs and tissues (15). The cellular and molecular reprogramming of lung cancer, including adenocarcinomas and NSCLC, has been identified in previous single-cell studies (16,17). Recently, Bischoff et al. (18) revealed the heterogeneous cellular composition of the tumor microenvironment across LUAD patients by single-cell-RNA sequencing. By combining single-cell and bulk RNA sequencing, some groups have developed a method for predicting the prognosis and immunotherapy responses of LUAD patients based on B cell (19) or natural-killer cell (20) genes. Although a recent study has shown that there is a the prognostic values of INTS7 for LUAD (21), indicated that INTS7 correlated with tumor microenvironment and immunotherapy responsiveness, the relationship of INTS7 for LUAD in single cell resolution and underlying mechanisms are poorly understood.
Using a public database cBioPortal (http://www.cbioportal.org/), we analyzed 4,541 tumor samples (from 4,391 patients) and found there is 4% overall mutation rate of INST7 in LAUD. Our research analyzed the expression of INTS7 in various cancers based on a pan-cancer database and a publicly available single-cell sequencing database. Additionally, protein-protein interaction (PPI) networks were constructed and functional annotations were undertaken for INTS7. We also selected the LUAD cell line H1299 for the knockdown experiments to further investigate the role of INTS7 in the biological behaviors of LUAD cells. Our results suggested that INTS7 was significantly overexpressed in LUAD, and the upregulation of INTS7 is associated with the risk factors of lung cancer metastasis and poor clinical characteristics. Thus, INTS7 could be a potential therapeutic target and a novel prognostic biomarker in LUAD. We present the following article in accordance with the MDAR reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1533/rc).
Methods
Single-cell-RNA sequencing data analysis
Single-cell-RNA sequencing data were collected from the Gene Expression Omnibus database (GSE131907). The cell annotation was quoted from the original text (16). The data were normalized, and the top 3,000 highly variable genes were selected and scaled. Next, raw single-cell–RNA sequencing data integration at different sample origins was performed with “FindIntegrationAnchors” and “IntegrateData” in the Seurat R package. A principle component analysis was then performed on certain selected highly variable genes, and the first 30 principle components with a resolution of 0.4 were used for cell clustering and mainly t-distributed stochastic neighbor embedding (t-SNE) visualization. The expression levels of INTS7 were visualized by “Dotplot” in the Seurat R package. The correlation of normalized gene expression to INTS7was calculated using the “Pearson” method, and the parameters were output by the “correlation” function.
Patients in The Cancer Genome Atlas (TCGA) database
Data from TCGA database were used to analyze INTS7 messenger RNA (mRNA) expression in LUAD patients and its relationship to the clinicopathological features, overall survival (OS), and disease-free survival (DFS) of the patients, (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga). The follow-up was completed on April 27, 2016, using TCGA database, and the data were analyzed using Xiantao (https://www.xiantao.love). TCGA is a cancer genomics program that is publicly available for any researcher to use. It has molecularly characterized >20,000 primary cancer types and matched normal samples covering 33 cancer types.
RNA-sequencing data of INTS7 in LUAD
The RNA-sequencing expression data of INTS7 in LUAD were downloaded from TCGA. In total, 535 LUAD and 59 normal tissue data items were obtained. The mRNA expression data are presented as the mean ± standard deviation (SD).
UALCAN and clinical proteomic tumor analysis
With the application of proteomic technologies, the Clinical Proteomic Tumor Analysis Consortium (CPTAC) was used to analyze the tumor samples by quantifying and identifying the protein, mass spectrometry results, and characterizing the proteome of each tumor biospecimen. Based on a comparison of the protein expressions of the tumor cells and normal cells, tumor-specific protein markers or specific antigens and drug targets were screened out, which provided insights into diagnosis. UALCAN (The University of ALabama at Birmingham CANcer data analysis) http://ualcan.path.uab.edu/analysis.html. is an easy to use, interactive web portal that is used to perform in-depth analyses of TCGA gene expression data. In this research, we used UALCAN to undertake a thorough analysis of INTS7 protein expression.
PPI networks and functional enrichment analysis
We used STRING (Search tool for the retrieval of interacting) https://string-db.org/ to search for the co-expression genes and construct the PPI networks with confidence score of >0.4. The results of the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Gene Ontology (GO) enrichment analyses of the co-expression genes were visualized using the “ggplot2” package.
Cell culture
The LUAD cell line (H1299) was obtained from the Shanghai Cell Bank at the Chinese Academy of Sciences. The cells were cultured in RPMI-1640 (Roswell Park Memorial Institute 1640, Biological Industries, Israel) with 10% fetal bovine serum (FBS). All the cells were maintained at 37 ℃ in a humidified incubator containing 5% carbon dioxide (CO2).
Cell transfection
The specific small-inhibitory RNA (siRNA) oligonucleotides targeting INTS7 were purchased from GenePharma (Suzhou, China), and the following sequences were selected as the interference targets: (I) GAGUUAGAGUCCUAACUAATT; and (II) CAGUAGACUUGAAGCUAAATT. Lipofectamine 2000 (Invitrogen, USA) was used to transfect the H1299 cells with 30 µM of siRNA in accordance with the manufacturer’s instructions. At 48 h post-transfection, the infected cells were examined for transfection efficiency by real-time quantitative reverse transcription and were used in the subsequent experiments.
Transwell migration and invasion assays
First, 24-well Transwell chambers (Corning, USA) were used to perform the migration and invasion assays. After being re-suspended with a serum-free medium, 6×104 cells were seeded into the upper chamber. The lower chamber was filled with 700 µL of the medium containing 10% FBS. After incubation for 48 h, the migrated cells were fixed on the filter inserts to stop the migration. The cells were stained with crystal violet (Thermo Scientific, USA) and then examined and photographed with an inverted microscope (Leica, USA). Subsequently, cellular migration was quantified by counting the number of cells from 10 random fields at ×100 magnification. Invasion assays were performed using the same procedures, except that the chamber was coated with 50 µL of matrix gel.
Colony formation assays and cell viability
For the colony formation assays, 1×103 siRNA-transfected cells/well were plated into 6-well plates at 37 ℃ in a humidified incubator containing 5% CO2. After 14 days, the medium was removed, and the cells were washed with phosphate buffered solution. The cells were then stained using crystal violet for 30 min. Finally, the colonies were photographed and counted. Cell proliferation and viability were detected using a Cell Counting Kit-8 (CCK-8; Dojindo, Japan). The transfected cells (2×103/well) were plated into 96-well plates (1,500 cells/well). At the indicated time (1, 2, 3, 4, 5, and 6 d), 10 µL of a CCK-8 solution was added into each well and incubated for 2 h. We measured the absorbance values at 450 nm with a microplate spectrometer.
Total RNA extraction and real-time PCR
Total RNA was extracted from the tissue samples or cell lines using the TRIzol reagent (Invitrogen, CA, USA), and the complementary deoxyribonucleic acid was synthesized in accordance with the manufacturer’s instructions. INTS7 expression was determined by real-time polymerase chain reaction (RT-PCR), which was performed using a Bio-Rad PCR instrument and the Takara SYBR Premix Extaq system (Takara Biotechnology, Dalian, China). The relative RNA expression of INTS7 was calculated using the 2−ΔΔCt method, with GAPDH (glyceraldehyde-3-phosphate dehydrogenase) as the normalization control. The primer sequences were as follows: INTS7: forward: TGGCACTGGAACAAGATG, reverse: CAACTACTGACTGGCTAAGA; GAPDH: forward: AATGGGCAGC CGTTAGGAAA, and reverse: GCGCCCAATACGACCA AATC.
Immunohistochemistry
In the present study, the patients’ samples were collected with the informed consent of all the patients upon approval by the Ethics Committee of The First Affiliated Hospital, School of Medical, Zhejiang University (No. 2021-735). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). First, 4-µm-thick sections of paraffin-embedded tissues were deparaffinized with xylene, rehydrated with gradient alcohol, and heated in citrate buffer for antigen retrieval. Next, the endogenous peroxidase activity was blocked by 5% bovine serum albumin (BSA) for 60 min at 25 ℃. After overnight incubation at 4 ℃ with the INTS7 primary polyclonal antibody (1:100, FineTest, Wuhan, China), the sections were incubated with the secondary antibody Horseradish Peroxidas Goat anti-rabbit IgG (1:100, A0277, Beyotime) for 90 min at room temperature. Finally, the sections were examined under light microscopy and evaluated using immuno-reactive scores.
Western blot analysis
Total proteins were extracted from cells and tissues with radioimmunoprecipitation assay lysis buffer mixed with phosphatase inhibitor tablets (Roche, 04906837001) and protease inhibitor tablet (Roche, 05892970001). Enhanced BCA Protein Assay Kit (Beyotime) was applied to evaluate the protein concentration and adjusted to equal concentration level according to the manufacturer’s instruction. Following the standard western blot procedure, the same amount of sample protein lysates was used to compare the expression of target proteins in different conditions. The primary antibodies used in this experiment were as followed: anti-GAPDH (1:1,000, GNI4310-GH, GNI, Japan), anti-INTS7 (1:1,000, FineTest, Wuhan, China). Then, the strips were incubated by secondary antibody. (Horseradish Peroxidas Goat anti-rabbit IgG 1:1,000, A0208, Beyotime) GAPDH was used to measure the relative abundance of proteins as an internal reference.
Flow cytometry
The H1299 cells were interfered with the siRNA control or INTS7 siRNA. Next, the cells were detached from the flasks using trypsin (GNM‑15050; GENOM, Ltd.), and the cell numbers were counted. The cells were then fixed with 4% paraformaldehyde for 15 min at room temperature. After washing with phosphate buffered saline (PBS), the cells were re‑suspended in 5% swine serum and blocked with an Fc receptor blocking solution (cat. No. 422301; Biolegend, Inc., San Diego, CA, USA) for 5 min. Aliquots of approximately 2×105 cells were then incubated with specific antibodies; that is, the annexin‑V‑Fluorescein Isothiocyanate antibody (cat. No. 70‑AP101‑100; Multiscience, Hangzhou, Zhejiang, China) and the immunoglobulin G (IgG) control, for 1 h at 4 ℃. The cells were then washed 3 times and analyzed by flow cytometry using the BD FACS Canto™ II system. A further analysis was conducted with the Flow Jo. Gating was set as <1% of the marker expression in the IgG control group.
Statistical analysis
All the statistical analyses were conducted using R (V 3.6.3), and the R package ggplot2 was used to visualize the differences. A Mann-Whitney U-test and a paired t-test were used to determine the difference between the LUAD and normal tissues and the experiment for INST7 siRNA inhibition in vitro. The chi-square test was used to examine the correlation between the clinicopathological variables and the INTS7 expression, and the survival analysis was performed using the Kaplan-Meier curves and a log-rank test. A P value <0.05 was considered statistically significant, all the experiment were repeated at least three times, for biological replicates.
Results
Single-cell transcriptional atlas of INTS7 in lung adenocarcinoma tissue
To investigate the mRNA expression pattern of INTS7 in multiple cancer types, we conducted a single-cell sequencing data analysis of a publicly available data set to compare INTS7 expression levels and percentages for different classifications of all kinds of cells (Figure 1A). First, we found that both the expression level and percentage of INTS7 were more increased in the malignant cells than the normal cells, which indicated that INTS7 may have the potential to be a prognostic marker. Next, we compared INTS7 expression in tissue levels and local tumor cells from primary sites. Specifically, we observed average expression of INTS7 in normal pulmonary tissues and normal lymph nodes at the early stage, advanced tumor cells from primary sites, pleural fluids, and metastatic lymph nodes at the local advanced stage, and brain metastasis at the distant metastasis stage (Figure 1B). The results showed that advanced tumor cells from primary sites and metastatic lymph nodes and metastatic adenocarcinoma cells had higher expression levels of INTS7 and were more abundant than normal cell types.
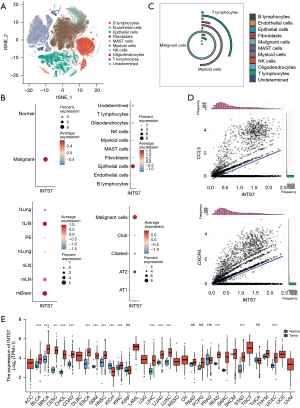
Generally, the tissue from a biopsy site contains various cell types (Figure 1C). Thus, we clustered and defined the cells and found that INTS7 was enriched in the epithelial cells. Further, we compared the normal epithelial subtypes to the malignant cells. We found that INTS7 was upregulated in process of the normal epithelial cell canceration and progress. Next, we selected INTS7 positive cells, and determined their relative counts (Figure 1D). In the gene expression matrix of INTS7 positive cells, the Pearson correlation analysis revealed the 4 genes with the highest positive correlations to INTS7. C-C motif chemokine 5 (CCL5) and C-X-C chemokine receptor type 4 (CXCR4) are both migration-associated genes. The overexpression of CXCR4 has also been linked to cancer metastasis, recurrence, and resistance to treatment in multiple cancer types (22). Further, research has shown that CXCR4 can recruit regulatory T cells to enhance tumor immune evasion (23). Similarly, CCL5 derived from myeloid-derived suppressor cells has been shown to enhance the migration and invasion of cancer cells in colon cancer (24). Thus, the migration ability of cancer cells is increased in those with high expression levels of INTS7 is increased. We also analyzed 33 cancer types by TCGA. As Figure 1E shows, INTS7 was significantly more upregulated in 31 of the 33 cancer type tissues than the adjacent normal tissues. Thus, the expression of INTS7 mRNA in different cancer types was found to be increased.
High INTS7 mRNA and protein levels are positively correlated with poor outcomes
As Figure 2A shows, an unpaired data analysis revealed that the mRNA expression levels of INTS7 in the adjacent normal tissues (n=59) was significantly lower than that in the LUAD tissues (n=535). To further assess INTS7 protein expression, we performed a comprehensive analysis of INTS7 protein expression using UALCAN based on the data obtained from CPTAC. The results indicated that the protein expression of INTS7 was higher in the LUAD tissues than the normal tissues (Figure 2B). Further, TCGA analysis showed that the expression of INTS7 differed between the Tumor classification stage I and Tumor classification stage II (P<0.05, Figure 2C). An immunohistochemical analysis of the LUAD tissues and the matched normal tissues revealed differences in the INTS7 expression levels, which was consistent with the results of the previous data analysis (Figure 2D). To examine whether INTS7 expression was associated with the OS and DFS rates, we divided TCGA cohorts into low- and high-expression subgroups based on the median expression of INTS7. The Kaplan-Meier analysis revealed that patients with low INTS7 expression levels had better OS and DFS rates than those with high INTS7 expression levels (Figure 2E). We also analyzed the relationship between INTS7 expression and the clinicopathological features of the LUAD patients and found statistically significant differences in terms of advanced Tumor classification stage, advanced Metastasis classification stage, and gender (Table 1). Additionally, similar as previously study (21), we conducted a univariate Cox analysis using data from TCGA database, and found that INTS7 was a predictor of OS (hazard ratio 1.496; 95% CI: 1.12–1.999; P=0.006) (Table S1).
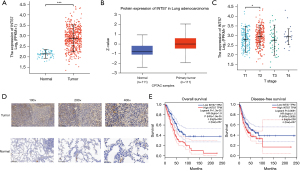
Table 1
| Characteristic | Low expression of INTS7 (n=267) |
High expression of INTS7 (n=268) |
P |
|---|---|---|---|
| T stage, n (%) | 0.031 | ||
| T1 | 99 (18.6) | 76 (14.3) | |
| T2 | 130 (24.4) | 159 (29.9) | |
| T3 | 29 (5.5) | 20 (3.8) | |
| T4 | 7 (1.3) | 12 (2.3) | |
| N stage, n (%) | 0.067 | ||
| N0 | 184 (35.5) | 164 (31.6) | |
| N1 | 40 (7.7) | 55 (10.6) | |
| N2 | 32 (6.2) | 42 (8.1) | |
| N3 | 2 (0.4) | 0 (0) | |
| M stage, n (%) | 0.009 | ||
| M0 | 177 (45.9) | 184 (47.7) | |
| M1 | 5 (1.3) | 20 (5.2) | |
| Gender, n (%) | < 0.001 | ||
| Female | 163 (30.5) | 123 (23) | |
| Male | 104 (19.4) | 145 (27.1) | |
| Race, n (%) | 0.876 | ||
| Asian | 3 (0.6) | 4 (0.9) | |
| Black or African American | 29 (6.2) | 26 (5.6) | |
| White | 203 (43.4) | 203 (43.4) | |
| Age, years, n (%) | 0.134 | ||
| ≤65 | 117 (22.7) | 138 (26.7) | |
| >65 | 138 (26.7) | 123 (23.8) | |
| OS event, n (%) | 0.016 | ||
| Alive | 185 (34.6) | 158 (29.5) | |
| Dead | 82 (15.3) | 110 (20.6) |
INTS7, integrator complex subunit 7; TCGA, The Cancer Genome Atlas; LUAD, lung adenocarcinoma; OS, overall survival.
PPI networks and functional annotations of INTS7
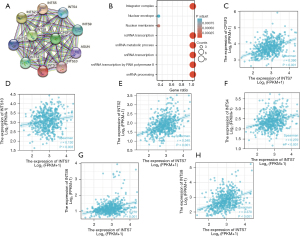
Data from the STRING database and KEGG and GO analyses were used to construct the PPI networks and perform the functional annotations. Figure 3A shows a PPI network for INTS7 and its 10 co-expression genes. Further, as Figure 3B shows, the functional enrichment analyses of the 11 involved genes revealed that the changes in the biological process of INTS7 were associated with non-coding RNA transcription, the snRNA metabolic process, snRNA transcription, snRNA transcription by RNA polymerase II, and snRNA processing. These genes appeared to be associated with a nuclear envelope based on the functional annotations. The expression correlation between INTS7 and its co-expressed genes in LUAD from TCGA were also analyzed (Figure 3C-3H).
Regulation of LUAD cell proliferation and invasion by INTS7
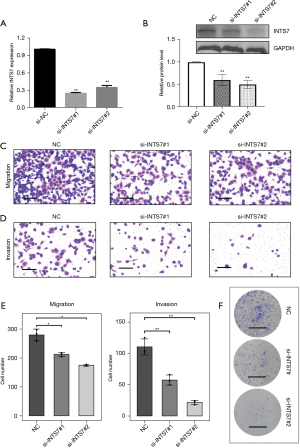
As a follow-up to our bioinformatic analysis, we explored the role of INTS7 in the biological behaviors of LUAD cells. The LUAD cell line H1299 was selected for the knockdown experiments. The H1299 cells were transfected with 2 siRNAs targeting INTS7. As Figure 4A,4B shows, we verified the transfection efficiency by PCR and Western blot. Notably, we found that H1299 cell migration (Figure 4C) and invasion (Figure 4D) were significantly impaired after INTS7 knockdown (Figure 4C-4E). We also found that silencing INTS7 repressed colony formation in the H1299 cells (Figure 4F).
INTS7 knockdown enhances the apoptosis and attenuates the proliferation capacity of LUAD cells
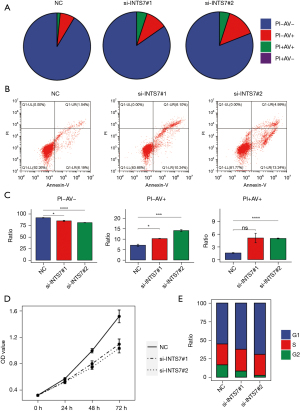
To investigate the effects of INTS7 knockdown on LUAD cell apoptosis, flow cytometry was used to detect Annexin V in the cells. A significant increase was observed for the mean apoptotic cell fraction (early apoptotic + late apoptotic) in the siINTS7 group compared to the siRNA control group, and by inhibiting INTS7, viable cell populations were reduced (Figure 5A-5C). Additionally, we used CCK-8 assays to investigate how INTS7 affects the proliferation viability of LUAD cells (Figure 5D), and found that the proliferation ability of the H1299 cells was significantly inhibited after INTS7 knockdown in a time-dependent manner. The percentage of proliferating cells in siINTS7 group was dramatically decreased, especially in G2 stage. (Figure 5E),
Discussion
LUAD is considered a heterogeneous disease. The survival time of LUAD patients differs depending on the pathological stages. A lack of reliable prognostic biomarkers and potential therapeutic targets make the prognosis and treatment of LUAD patients difficult (25-27). The function of INTS7 has been reported in some studies (13,14); however, the relevance of INTS7 to the diagnosis and prognosis of LUAD patients had not previously been investigated. In our study, we focused on INTS7 and investigated the clinicopathological significance of INTS7 in LUAD patients.
INTS7 encodes a subunit of the integrator complex, which mediates the processing of snRNAs and is associated with RNA polymerase II. Thus, it is essential to investigate the prognostic value of INTS7 in LUAD patients. In our study, we conducted a pan-cancer analysis and found that the mRNA expression of INTS7 was aberrantly expressed in multiple cancer types. Further, we confirmed that the expression level of INTS7 was more increased in LUAD tissues than normal tissues. By analyzing data from TCGA database, we found that INTS7 played a critical role in the progression of LUAD and was associated with an advanced T stage, advanced M stage, and gender. In addition, the Kaplan-Meier analysis showed that the OS and DFS rates of LUAD patients were inversely correlated with INTS7 expression. Based on the findings from TCGA, we further knocked down the expression of INTS7 in the LUAD cells. We found that silencing the expression of INTS7 inhibited the proliferation of LUAD cells. In addition, the depletion of INTS7 inhibited the migration and invasion of LUAD cells. These results revealed that INTS7 could serve as a potential prognostic biomarker to identify LUAD patients with poor clinical outcomes.
Conclusions
In summary, in this research, we demonstrated that INTS7 was upregulated in the LUAD tissues and had an inverse relationship with the OS and DFS rates of LUAD patients. Knocking down INTS7 inhibited LUAD cell viability and invasion in vitro. Our results indicate that INTS7 could serve as a promising therapeutic target and a novel prognostic biomarker in LUAD.
Acknowledgments
We thank Alibaba-Zhejiang University Joint Research Center of Future Digital Healthcare and Alibaba Cloud for digital cloud storage support to the study, and we also thank Professor Ting Chen in Department of Cardiology, the First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, China for the funding support.
Funding: This work was supported by a grant from the National Natural Science Foundation of China (No. 82170489).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1533/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1533/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1533/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This research was approved by the Ethics Committee of The First Affiliated Hospital, School of Medical, Zhejiang University (No. 2021-735). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was obtained from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Brustugun OT, Grønberg BH, Fjellbirkeland L, et al. Substantial nation-wide improvement in lung cancer relative survival in Norway from 2000 to 2016. Lung Cancer 2018;122:138-45. [Crossref] [PubMed]
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553:446-54. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Pore MM, Hiltermann TJ, Kruyt FA. Targeting apoptosis pathways in lung cancer. Cancer Lett 2013;332:359-68. [Crossref] [PubMed]
- Yuan M, Huang LL, Chen JH, et al. The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Signal Transduct Target Ther 2019;4:61. [Crossref] [PubMed]
- Attarian S, Rahman N, Halmos B. Emerging uses of biomarkers in lung cancer management: molecular mechanisms of resistance. Ann Transl Med 2017;5:377. [Crossref] [PubMed]
- Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet 2017;389:299-311. [Crossref] [PubMed]
- Wu Y, Wei J, Zhang W, et al. Serum Exosomal miR-1290 is a Potential Biomarker for Lung Adenocarcinoma. Onco Targets Ther 2020;13:7809-18. [Crossref] [PubMed]
- Singh SS, Dahal A, Shrestha L, et al. Genotype Driven Therapy for Non-Small Cell Lung Cancer: Resistance, Pan Inhibitors and Immunotherapy. Curr Med Chem 2020;27:5274-316. [Crossref] [PubMed]
- Wu K, House L, Liu W, et al. Personalized targeted therapy for lung cancer. Int J Mol Sci 2012;13:11471-96. [Crossref] [PubMed]
- Federico A, Rienzo M, Abbondanza C, et al. Pan-Cancer Mutational and Transcriptional Analysis of the Integrator Complex. Int J Mol Sci 2017;18:936. [Crossref] [PubMed]
- Baillat D, Wagner EJ. Integrator: surprisingly diverse functions in gene expression. Trends Biochem Sci 2015;40:257-64. [Crossref] [PubMed]
- Rutkowski RJ, Warren WD. Phenotypic analysis of deflated/Ints7 function in Drosophila development. Dev Dyn 2009;238:1131-9. [Crossref] [PubMed]
- Nakagawa H, Tategu M, Yamauchi R, et al. Transcriptional regulation of an evolutionary conserved intergenic region of CDT2-INTS7. PLoS One 2008;3:e1484. [Crossref] [PubMed]
- Qiu X, Mao Q, Tang Y, et al. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 2017;14:979-82. [Crossref] [PubMed]
- Kim N, Kim HK, Lee K, et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat Commun 2020;11:2285. [Crossref] [PubMed]
- Guo X, Zhang Y, Zheng L, et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat Med 2018;24:978-85. [Crossref] [PubMed]
- Bischoff P, Trinks A, Obermayer B, et al. Single-cell RNA sequencing reveals distinct tumor microenvironmental patterns in lung adenocarcinoma. Oncogene 2021;40:6748-58. [Crossref] [PubMed]
- Song P, Li W, Wu X, et al. Integrated analysis of single-cell and bulk RNA-sequencing identifies a signature based on B cell marker genes to predict prognosis and immunotherapy response in lung adenocarcinoma. Cancer Immunol Immunother 2022;71:2341-54. [Crossref] [PubMed]
- Song P, Li W, Guo L, et al. Identification and Validation of a Novel Signature Based on NK Cell Marker Genes to Predict Prognosis and Immunotherapy Response in Lung Adenocarcinoma by Integrated Analysis of Single-Cell and Bulk RNA-Sequencing. Front Immunol 2022;13:850745. [Crossref] [PubMed]
- Li X, Yao Y, Qian J, Jin G, Zeng G, Zhao H. Overexpression and diagnostic significance of INTS7 in lung adenocarcinoma and its effects on tumor microenvironment. Int Immunopharmacol. 2021 Dec;101(Pt B):108346.
- Chatterjee S, Behnam Azad B, Nimmagadda S. The intricate role of CXCR4 in cancer. Adv Cancer Res 2014;124:31-82. [Crossref] [PubMed]
- Gil M, Komorowski MP, Seshadri M, et al. CXCL12/CXCR4 blockade by oncolytic virotherapy inhibits ovarian cancer growth by decreasing immunosuppression and targeting cancer-initiating cells. J Immunol 2014;193:5327-37. [Crossref] [PubMed]
- Kan JY, Wu DC, Yu FJ, et al. Chemokine (C-C Motif) Ligand 5 is Involved in Tumor-Associated Dendritic Cell-Mediated Colon Cancer Progression Through Non-Coding RNA MALAT-1. J Cell Physiol 2015;230:1883-94. [Crossref] [PubMed]
- Rami-Porta R, Bolejack V, Crowley J, et al. The IASLC Lung Cancer Staging Project: Proposals for the Revisions of the T Descriptors in the Forthcoming Eighth Edition of the TNM Classification for Lung Cancer. J Thorac Oncol 2015;10:990-1003.
- Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol 2015;10:1243-60. [Crossref] [PubMed]
- Torre LA, Siegel RL, Jemal A. Lung Cancer Statistics. Adv Exp Med Biol 2016;893:1-19. [Crossref] [PubMed]


