Prognostic significance of FoxM1 expression in non-small cell lung cancer
Introduction
Lung cancer is a leading cause of cancer deaths in the world. Approximately 80–85% of all lung cancers are non-small-cell lung cancer (NSCLC) (1). The prognosis for lung cancer patients is generally poor, with an overall 5-year survival rate of approximately 15%, and the 5-year overall survival (OS) rate for patients with NSCLC has not been markedly improved (2,3). Several independent prognostic factors for survival have been identified: performance status (PS), disease stage, age, sex and amount of weight lost (4). However, the discriminant value of most potential prognostic biological markers is insufficient to predict the optimal therapeutic course for an individual (5).
FoxM1 is a member of the forkhead transcription factor family, which plays an important role in controlling the cell cycle (6,7). The transcription factor FoxM1plays a vital role in the regulation of a wide range of biological processes, including cell cycle progression, cell proliferation, cell differentiation, angiogenesis, apoptosis, DNA damage repair, and tissue homeostasis (8). Previous studies have demonstrated that overexpression of FoxM1 is associated with tumorigenesis and progression in a variety of human cancers, including the breast, colorectum, lung, prostate, liver, pancreas, cervix, blood, and nervous system (9-15). Thus, it is acting with oncogenic properties.
Many studies have evaluated whether the overexpression of FoxM1 may be a prognostic factor for survival in patients with NSCLC. However, the results of the studies are inconclusive and no consensus has been reached. Thus, we conducted a meta-analysis of all available studies relating FoxM1 with the clinical outcome in patients with lung cancer.
Methods
Search strategy and study selection
The electronic databases PubMed was searched for studies to include in the present meta-analysis. An upper date limit of Jan 01, 2015 was applied; we used no lower date limit. Searches included the terms “lung”, “cancer or carcinoma or tumor or neoplasm”, “FoxM1”, “FoxM1a” or “FoxM1b” or “FoxM1c” and “prognosis”. We also reviewed the Cochrane Library for relevant articles. The references reported in the identified studies were also used to complete the search.
Studies eligible for inclusion in this meta-analysis met the following criteria: (I) measure FoxM1 expression in the primary lung cancer with IHC (immunohistochemistry); (II) provide information on survival (i.e., OS, studies investigating response rates only were excluded); (III) when the same author reported results obtained from the same patient population in more than one publication, only the most recent report, or the most complete one, was included in the analysis. Two authors independently determined study eligibility. Disagreements were resolved by consensus.
Data extraction and quality assessment
The final articles included were assessed independently by two reviewers. Data retrieved from the reports included author, publication year, patient source, histology, study design, test method, positive, follow-up and survival data (Table 1). If data from any of the above categories were not reported in the primary study, items were treated as “not applicable”. We did no contact the author of the primary study to request the information. We did not use prespecified quality-related inclusion or exclusion criteria and did not weigh each study by a quality score, because the quality score has not received general agreement for use in a meta-analysis, especially observational studies (16).
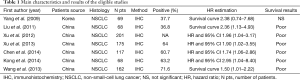
Full table
Statistical methods
For the quantitative aggregation of the survival results, we measured the impact of FoxM1 overexpression on survival by HR between the two survival distributions. HRs and 95% confidence intervals (CIs) were used to combine as the effective value. If the HRs and their 95% CIs were given explicitly in the articles, we used crude ones. When these variables were not given explicitly, they were calculated from the available numerical data using methods reported by Parmar et al. (17).
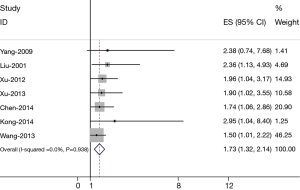
Heterogeneity of the individual HRs was calculated with χ2 tests according to Peto’s method (18). Heterogeneity test with inconsistency index (Ι2) statistic and Q statistic was performed. If HRs were found to have fine homogeneity, a fixed effect model was used for secondary analysis; if not, a random-effect model was used. DerSimonian-Laird random effects analysis (19) was used to estimate the effect of FoxM1 overexpression on survival. By convention, an observed HR >1 implies worse survival for the group with FoxM1 overexpression. The impact of FoxM1 on survival was considered to be statistically significant if the 95% confidence interval (CI) did not overlap with 1. Horizontal lines represent 95% CIs. Each box represents the HR point estimate, and its area is proportional to the weight of the study. The diamond (and broken line) represents the overall summary estimate, with CI represented by its width. The unbroken vertical line is set at the null value (HR =1.0). Evidence of publication bias was sought using the methods of Egger et al. (20) and of Begg et al. (21). Intercept significance was determined by the t-test suggested by Egger (P<0.05 was considered representative of statistically significant publication bias). All of the calculations were performed by STATA version 11.0 (Stata Corporation, College Station, TX, USA).
Results
Study selection and characteristics
Seven studies (22-28) published between 2009 and 2014 were eligible for this meta-analysis. All reported the prognostic value of FoxM1 status for survival in lung cancer patients. The total number of patients included was 860, ranging from 68 to 201 patients per study (median 122). The major characteristics of the 7 eligible publications are reported in Table 1. The studies were conducted in two countries (China and Korea). Among the seven studies, all studies were performed in Asian populations. All patients in the eligible studies were determined by pathological stage.
All of the studies reported the prognostic value of FoxM1 status for survival in patients with lung cancer. Four of the seven studies reported HRs (multivariate analysis) explicitly. A HR on OS could be extracted for three studies. Six of the seven studies identified FoxM1 overexpression as an indicator of poor OS, and the other one study showed no statistically significant impact of FoxM1 overexpression on OS.
Meta-analysis
The results of the meta-analysis were shown in Figure 1. Overall, the combined HR for all 7 eligible studies evaluating FoxM1 overexpression on OS was 1.73 (1.32–2.14), suggesting that FoxM1 overexpression was associated with poor prognosis for lung cancer. No significant heterogeneity was observed among the studies (Q=12.39, I2=36.9%, P=0.938).
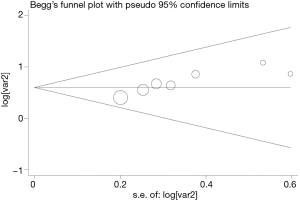
Publication bias
Begg’s funnel plot and Egger’s test were performed to assess the publication bias in the literature. All seven eligible studies investigating FoxM1 overexpression on OS yielded a Begg’s test score of P=0.420 and an Egger’s test score of P=0.251, meanwhile according to the funnel plot (Figure 2), the absence of publication bias was found. These results suggested that there were no publication biases in these subgroup analyses.
Discussion
FoxM1, which consists of more than 50 amino acid residues and is characterized by a conserved 100 amino acid DNA binding domain, is a member of the FoxM1 family; FoxM1 is also a transcription factor that plays important roles in cell proliferation, organogenesis, aging and cancer (6,29,30). FoxM1 controls mitotic entry through the periodic upregulation of a group of genes that are maximally expressed as cells progress through late G2 and into M phase (6). Two of its target genes are CCNB1 and PLK1, and these form part of a kinase-driven positive feedback loop that leads to the phosphorylation of FoxM1 and potentiation of its activity (31,32).
Our present meta-analysis is the first to evaluate the correlation between FoxM1 overexpression and survival in patients with NSCLC. The meta-analysis combined 7 publications including 860 patients with lung cancer to yield statistics, indicating statistically significant role of FoxM1 on OS in NSCLC. Combined hazard ratios suggested that FoxM1 overexpression was associated with poor prognosis of OS (HR =1.73, 95% CI: 1.32–2.14) in patients with NSCLC.
The heterogeneity issue was complicated in the systematic review and meta-analysis was. We found no significant heterogeneity among all studies included and subgroup analysis. Another potential source of bias is related to the method of HR and 95% CI extrapolation. If these statistics were not reported by the authors, we calculated them from the data available in the article. If this was not possible, we extrapolated them from the survival curves, necessarily making assumptions about the censoring process. Data for multivariate survival analysis reported in the article were included in the present systematic review with meta-analysis; if these data were not available, data calculated from survival curves by univariate analysis were included. These results should be confirmed by an adequately designed prospective study. Furthermore, the exact value of FoxM1 overexpression status needs to be determined by appropriate multivariate analysis. Unfortunately, few prospectively designed prognostic studies concerning biomarkers have been reported; thus, our collection of many retrospective studies revealed more significance.
Publication bias (33) is a major concern for all forms of meta-analysis; positive results tend to be accepted by journals, while negative results are often rejected or not even submitted. The present analysis does not support publication bias; the obtained summary statistics likely approximate the actual average. However, it should be noted that our meta-analysis could not completely exclude biases. For example, the study was restricted to papers published in English and Chinese, which probably introduced bias.
In conclusion, despite the limitations described above, our meta-analysis is the first study to systematically estimate the association between FoxM1 expression detected by IHC and lung cancer survival. As determined in our meta-analysis, we concluded that FoxM1 overexpression was associated with poor OS in lung cancer. To strengthen our findings, well-designed prospective studies with better standardized assessment of prognostic markers should help to explore the relation between FoxM1 overexpression and survival of lung cancer.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. CA Cancer J Clin 2014;64:9-29. [Crossref] [PubMed]
- Bernard ME, Clump DA, Lalonde R, et al. Radiation therapy for locally advanced lung cancer. Transl Cancer Res 2015;4:356-71.
- Agalioti T, Giannou AD, Stathopoulos GT. Pleural involvement in lung cancer. J Thorac Dis 2015;7:1021-30. [PubMed]
- Wang LB, Chuang EY, Lu TP. Identification of predictive biomarkers for ZD-6474 in lung cancer. Transl Cancer Res 2015;4:324-31.
- Sahiner I, Vural GU. Positron emission tomography/computerized tomography in lung cancer. Quant Imaging Med Surg 2014;4:195-206. [PubMed]
- Laoukili J, Stahl M, Medema RH. FoxM1: at the crossroads of ageing and cancer. Biochim Biophys Acta 2007;1775:92-102.
- Koo CY, Muir KW, Lam EW. FOXM1: From cancer initiation to progression and treatment. Biochim Biophys Acta 2012;1819:28-37.
- Teh MT, Wong ST, Neill GW, et al. FOXM1 is a downstream target of Gli1 in basal cell carcinomas. Cancer Res 2002;62:4773-80. [PubMed]
- Kim IM, Ackerson T, Ramakrishna S, et al. The Forkhead Box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer. Cancer Res 2006;66:2153-61. [Crossref] [PubMed]
- Ahmad A, Wang Z, Kong D, et al. FoxM1 down-regulation leads to inhibition of proliferation, migration and invasion of breast cancer cells through the modulation of extra-cellular matrix degrading factors. Breast Cancer Res Treat 2010;122:337-46. [Crossref] [PubMed]
- Nakamura S, Hirano I, Okinaka K, et al. The FOXM1 transcriptional factor promotes the proliferation of leukemia cells through modulation of cell cycle progression in acute myeloid leukemia. Carcinogenesis 2010;31:2012-21. [Crossref] [PubMed]
- Priller M, Pöschl J, Abrão L, et al. Expression of FoxM1 is required for the proliferation of medulloblastoma cells and indicates worse survival of patients. Clin Cancer Res 2011;17:6791-801. [Crossref] [PubMed]
- Chu XY, Zhu ZM, Chen LB, et al. FOXM1 expression correlates with tumor invasion and a poor prognosis of colorectal cancer. Acta Histochem 2012;114:755-62. [Crossref] [PubMed]
- Huang C, Qiu Z, Wang L, et al. A novel FoxM1-caveolin signaling pathway promotes pancreatic cancer invasion and metastasis. Cancer Res 2012;72:655-65. [Crossref] [PubMed]
- Okada K, Fujiwara Y, Takahashi T, et al. Overexpression of forkhead box M1 transcription factor (FOXM1) is a potential prognostic marker and enhances chemoresistance for docetaxel in gastric cancer. Ann Surg Oncol 2013;20:1035-43. [Crossref] [PubMed]
- Altman DG. Systematic reviews of evaluations of prognostic variables. BMJ 2001;323:224-8. [Crossref] [PubMed]
- Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med 1998;17:2815-34. [Crossref] [PubMed]
- Yusuf S, Peto R, Lewis J, et al. Beta blockade during and after myocardial infarction: an overview of the randomized trials. Prog Cardiovasc Dis 1985;27:335-71. [Crossref] [PubMed]
- DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials 1986;7:177-88. [Crossref] [PubMed]
- Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997;315:629-34. [Crossref] [PubMed]
- Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics 1994;50:1088-101. [Crossref] [PubMed]
- Yang DK, Son CH, Lee SK, et al. Forkhead box M1 expression in pulmonary squamous cell carcinoma: correlation with clinicopathologic features and its prognostic significance. Hum Pathol 2009;40:464-70. [Crossref] [PubMed]
- Liu YQ, Guo RH, Liu LK, et al. Correlation between expression of forkhead box M1 (FOXM1) and clinicopathological features and prognosis in patients with non-small cell lung cancer (NSCLC). Zhonghua Zhong Liu Za Zhi 2011;33:426-30. [PubMed]
- Xu N, Wu SD, Wang H, et al. Involvement of FoxM1 in non-small cell lung cancer recurrence. Asian Pac J Cancer Prev 2012;13:4739-43. [Crossref] [PubMed]
- Xu N, Jia D, Chen W, et al. FoxM1 is associated with poor prognosis of non-small cell lung cancer patients through promoting tumor metastasis. PLoS One 2013;8:e59412. [Crossref] [PubMed]
- Chen PM, Cheng YW, Wang YC, et al. Up-regulation of FOXM1 by E6 oncoprotein through the MZF1/NKX2-1 axis is required for human papillomavirus-associated tumorigenesis. Neoplasia 2014;16:961-71. [Crossref] [PubMed]
- Kong FF, Qu ZQ, Yuan HH, et al. Overexpression of FOXM1 is associated with EMT and is a predictor of poor prognosis in non-small cell lung cancer. Oncol Rep 2014;31:2660-8. [PubMed]
- Wang Y, Wen L, Zhao SH, et al. FoxM1 expression is significantly associated with cisplatin-based chemotherapy resistance and poor prognosis in advanced non-small cell lung cancer patients. Lung Cancer 2013;79:173-9. [Crossref] [PubMed]
- Carlsson P, Mahlapuu M. Forkhead transcription factors: key players in development and metabolism. Dev Biol 2002;250:1-23. [Crossref] [PubMed]
- Kaestner KH, Knochel W, Martinez DE. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev 2000;14:142-6. [PubMed]
- Major ML, Lepe R, Costa RH. Forkhead box M1B transcriptional activity requires binding of Cdk-cyclin complexes for phosphorylation-dependent recruitment of p300/CBP coactivators. Mol Cell Biol 2004;24:2649-61. [Crossref] [PubMed]
- Fu Z, Malureanu L, Huang J, et al. Plk1-dependent phosphorylation of FoxM1 regulates a transcriptional programme required for mitotic progression. Nat Cell Biol 2008;10:1076-82. [Crossref] [PubMed]
- Begg CB, Berlin JA. Publication bias(a problem in interpreting medical data). J R Stat Soc A 1988;151:419-63. [Crossref]


