
Experimental study on the suppressive effect of B3GNT3 on the apoptosis of lung adenocarcinoma cells and its application in early screening for lung adenocarcinoma
Highlight box
Key findings
• High expression of the secreted protein B3GNT3 in lung adenocarcinoma is closely related to prognosis.
What is known and what is new?
• Serological tumor markers have been widely promoted and applied in the clinic for liver cancer, colorectal cancer, mammary cancer and other diseases.
• B3GNT3 is the key upstream regulator of PD-L1.
What is the implication, and what should change now?
• B3GNT3 can serve as a potential biological marker for the early screening of lung adenocarcinoma.
Introduction
The incidence and mortality rates of lung cancer rank first among those of all malignant tumors, posing a serious public health problem. There are approximately 1.2 million patients with lung cancer worldwide, and those dying of lung cancer account for 25% of those dying of malignant tumors. In recent years, the proportion of lung adenocarcinoma has gradually increased in lung cancer (1). Non-small cell lung cancer (NSCLC) accounts for 80% of total lung cancer cases. As the most common histological type of NSCLC, lung adenocarcinoma is more common in elderly patients, with characteristics of early metastasis, rapid progression, high incidence rate and high mortality rate (2,3). Most patients with lung adenocarcinoma are diagnosed at an advanced stage (4). At present, the treatments for lung adenocarcinoma mainly include surgery, radiotherapy and chemotherapy. Although the above treatments can reduce the risk of death in patients with lung adenocarcinoma to a certain extent, the overall survival of patients with lung adenocarcinoma is still very low, and the prognosis is poor because it is difficult to find and easily recurs (5). Therefore, clarifying the molecular mechanisms of lung adenocarcinoma and identifying novel candidate biomarkers are prerequisites for the early diagnosis of lung adenocarcinoma, which is a key factor for effective treatment.
Recently, many scholars have tried to achieve early diagnosis in patients with lung adenocarcinoma by screening for effective biological markers (6). Tumor markers refer to a class of substances abnormally synthesized and released by malignant tumor cells or a class of substances that increase the host’s response to tumor stimulation, existing in tumor cells or hosts’ tissues, body fluids or excrement in the forms of enzymes, hormones, antigen or oncogene products (7,8). Serum tumor markers have the advantages of corresponding high specificity, high sensitivity, convenience, ease of access, and minimal trauma, and research on tumor markers has also been ongoing for a long time (9). Serological tumor markers [alpha fetoprotein (AFP), carcinoembryonic antigen (CEA), etc.] have been widely promoted and applied in the clinic for liver cancer, colorectal cancer, mammary cancer and other diseases (10). However, effective serum tumor markers have not been screened for lung adenocarcinoma.
With profound research on genomics, numerous researchers have attempted to obtain specific tumor markers through bioinformatic analysis of databases such as The Cancer Genome Atlas (TCGA) database and Gene Expression Omnibus (GEO) database and by screening of long noncoding RNAs (lncRNAs) and microRNAs (miRNAs) in lung cancer (11-13). However, comprehensive specific serum tumor marker screening for lung adenocarcinoma via TCGA database analysis has not been performed.
On this basis, this study preliminarily screened specific serum tumor markers in patients with lung adenocarcinoma through TCGA database analysis. The sensitivity and specificity of the identified marker were defined through validation in patients with lung adenocarcinoma. Moreover, lung adenocarcinoma cell lines were cultured in vitro, and the specific molecular regulatory mechanism of this tumor marker for lung adenocarcinoma was finally defined by molecular biological techniques. On the basis of previous studies, this study further explored the significance of beta-1,3-N-acetylglucosaminyltransferase 3 (B3GNT3) in early screening of lung adenocarcinoma and its specific molecular regulation mechanism for the occurrence and development of lung adenocarcinoma. Finally, it is clear that B3GNT3 is the key upstream regulator of programmed cell death-ligand 1 (PD-L1). We present the following article in accordance with the MDAR reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1175/rc).
Methods
Data source and analysis
The TCGA database was searched for tumor samples of patients with lung adenocarcinoma and normal control samples, and differentially expressed genes were defined as those with |log2fold change (FC)| >2 and P.adj <0.05 according to the DESeq2 software package in R 4.0.3. A total of 3,612 detectable secreted proteins in peripheral blood were obtained through the MetaSecKB database. MetazSecKB is a secretome and subcellular proteome knowledgebase specifically designed for metazoan, i.e., human and animals. According to the P.adj and |log2FC|, volcano plots were drawn to visualize the expression of secreted proteins. Finally, the secreted proteins closely related to the occurrence and development of lung cancer were screened out.
Clinical sample source
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Qiqihar Medical University (No. 20200425). Informed consent was taken from all individual patients. Data for patients with lung adenocarcinoma treated surgically in our hospital from January 2018 to January 2019 were collected retrospectively according to the inclusion and exclusion criteria. The inclusion criteria were as follows: (I) patients first diagnosed with lung adenocarcinoma in our hospital and (II) patients who did not undergo radiotherapy, chemotherapy or immunotherapy before surgery. The exclusion criteria were as follows: (I) patients with multiple tumors; (II) patients who underwent radiotherapy, chemotherapy and immunotherapy before surgery; and (III) patients with a history of severe liver and renal dysfunction and mental illness. The same number of healthy people who had received physical examinations from hospitals were obtained by propensity ratio analysis and assigned to the normal control group. Peripheral blood samples were collected from the control and lung adenocarcinoma groups 1 week before surgery and from patients with lung adenocarcinoma 1 week after surgery.
The diagnostic criteria for lung adenocarcinoma was as follows: all patients were diagnosed with lung adenocarcinoma through chest computed tomography (CT), sputum exfoliative cytology examination, fiberoptic bronchoscope, lung puncture or surgical pathological examination, with clinical stages of Ia–IVb.
Cell culture
The human normal lung epithelial cell line BEAS-2B and four human lung adenocarcinoma cell lines (A549, H1975, Calu3 and H1395) were purchased from American Type Culture Collection (ATCC). According to the criteria provided by ATCC for cell culture, the normal lung epithelial cell line BEAS-2B was cultured with BEGM medium CC-3170 (Lonza/Clonetics, CC-3170). The four human lung adenocarcinoma cell lines were cultured with RPMI medium (Gibco, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco), 100 U/mL penicillin and 100 mg/L streptomycin. The cells were placed in an incubator at 37 ℃ with 5% CO2 for culture.
Quantitative polymerase chain reaction (qPCR)
Total RNA in tissues and cells was extracted with TRIzol reagent, which was reverse transcribed into cDNA with the RNA reverse transcription kit, 5X All-In-One RT MasterMix (ABM, Cat. No. G492, China), taking complementary DNA (cDNA) as a template for qPCR with EvaGreen 2X qPCR MasterMix (ABM, Cat. MasterMix-S). The B3GNT3 forward primer sequence was CAGCACGTTCAGAACTTCCTC, the reverse primer sequence was GCGCACATAGTTGCTAGGGG, and the results were analyzed by relative quantitative analysis.
Enzyme-linked immunosorbent assay (ELISA)
Fresh blood from patients with tumors was placed on ice for half an hour and centrifuged at 15,000 rpm at 4 ℃ for 30 min. The upper serum level was taken and processed with a human B3GNT3 ELISA kit (Sai Berry, Cat. SEJ574Hu02, China) to determine B3GNT3 expression in serum.
Short hairpin RNA (shRNA) constructs and transfection
shRNA plasmids (Hanheng Biotechnology, China) with the target sequence CCGCTTCCTACCTTATGAGAT were constructed for B3GNT3 gene expression interference and packaged as lentivirus (with a titer of 810 IU/mL). The cells were spread in 6-well plates. When the confluence of the cells reached 50–70%, 20 µL shRNA was added to each well, the transfection efficiency was observed by microscopy after 48 h, and the transfected cells were screened with 2 µg/mL puromycin. After 5 days of screening, the medium was replaced with normal medium and subamplified to a confluence of 70% for subsequent experiments. The efficiency of B3GNT3 knockdown was determined by qRT-PCR and Western blotting (WB).
WB
The cells of each group were collected with efficient RIPA lysis buffer (Solarbio, Cat. R0020, China), lysed at 4 ℃, and then centrifuged at 15,000 rpm for 15 min for protein extraction. The protein was quantified with a bicinchoninic acid (BCA) protein quantification kit (Thermo Fisher, Cat. 23235, USA). After the quantification of the protein concentration, sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) electrophoresis was conducted for each tissue/cell. Subsequently, the proteins separated by electrophoresis were transferred to a polyvinylidene fluoride (PVDF) membrane with a transfer device; the membrane was then blocked with 5% skim milk for 1 h and rinsed in tris-buffered saline with Tween 20 (TBST). Primary antibodies against the target proteins were then added for incubation at 4 ℃ overnight, and then a secondary antibody was added for incubation for 1 hour. Finally, the PVDF membrane was exposed to test through a chemiluminescence imaging system.
In this study, rabbit anti-B3GNT3 antibody (Abcam, Cat. ab70156, UK) was adopted as the B3GNT3 antibody with a dilution ratio of 1:1,000; rabbit anti-β-actin antibody (Abcam, Cat. 8227) was used as the internal reference antibody with a dilution ratio of 1:1,000; rabbit anti-Bax antibody (Abcam, ab32503) was used as the BAX antibody with a dilution ratio of 1:2,000; rabbit anti-Bcl2 antibody (Abcam, ab32124) was used as the Bcl2 antibody; and HRP anti-rabbit IgG antibody (Abcam, ab288151) was used as the secondary antibody.
Cell Counting Kit-8 (CCK-8) proliferation assay
Lenti-shB3GNT3 cells and the control cells were inoculated in 96-well plates at 1,000 cells/well and cultured for 24, 48, 72 and 96 h. CCK-8 detection reagent was added at 1:10 (Vazyme Biotech, Cat. A311-02, China), and the optical density (OD) was detected at 450 nm.
Apoptosis detected by ELISA
Apoptosis was detected by an ELISA apoptosis kit (meeck, cat. APOAC-1KT, USA). After A549 cells were treated under different grouping conditions, the cells were first collected by centrifugation and then resuspended in 200 µL of cytolytic buffer, followed by quiescence for 30 min. After centrifugation at 1,000 r/min for 10 min, 20 µL of the supernatant was removed and added to the ELISA plate. Then, 80 µL of immunoreactive reagent was added to each well of the plates and incubated for 2 h at room temperature. The supernatant was removed, and the well was rinsed three times. Then, 100 µL of buffer was added and incubated for 30 min. The OD was measured at a wavelength of 405 nm, with the substrate buffer as the blank control group.
Statistical analysis
Statistical analyses of the data used were performed in SPSS 24.0 (IBM, New York, USA) software. For the count variables, the chi-square test was used. For measurement data the independent samples t-test was used for those that obeyed normal distribution, and the results were expressed as mean ± standard deviation; for those that did not obey normal distribution the Mann-Whitney U test was used, and the results were expressed as median [quartiles]. P<0.05 indicates a statistical difference.
Results
Bioinformatics analysis revealed that B3GNT3 is a key secreted protein of potential blood biomarkers for lung cancer
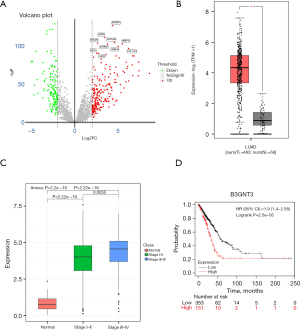
A total of 535 tumor samples and 59 normal control samples were obtained after searching the TCGA database. A total of 209 up-regulated genes and 140 down-regulated genes were set as differentially expressed genes from these samples. The differentially expressed genes acquired from the TCGA database were cross-compared with the MetaSecKB database, and 3,612 detectable secreted proteins in peripheral blood were obtained. Screening conditions were set for the differentially expressed secreted proteins (|log2FC| >2, P.adj <0.05), and the variability of each secreted protein is shown by volcano plots (Figure 1A). The results showed that B3GNT3 (logFC =4.381387, P<0.01) was the secreted protein with the most significant differential expression. The analysis of 535 samples from patients with cancers and 59 samples from normal controls in TCGA showed significantly high B3GNT3 expression in samples from patients with lung adenocarcinoma (Figure 1B). Further detection of B3GNT3 expression in each sample from patients with lung adenocarcinoma according to the clinical stage of patients with tumors showed that the higher the clinical stage was, the higher the B3GNT3 expression in patients with lung adenocarcinoma (Figure 1C, P<0.05), suggesting a gradual increase in B3GNT3 expression during disease progression in patients with lung adenocarcinoma. Patients with lung adenocarcinoma were divided into high and low expression groups according to the median B3GNT3 expression, and Kaplan-Meier (K-M) curves were drawn to assess the survival of both groups, which showed that the survival cycle of the high B3GNT3 expression group was significantly worse than that of the low B3GNT3 expression group (Figure 1D). The results of the above bioinformatics analysis showed that B3GNT3 is closely associated with tumor grade in patients with lung adenocarcinoma, and the prognosis of patients with high B3GNT3 expression was significantly worse. Therefore, B3GNT3 may serve as a key secreted protein and potential blood biomarker in lung cancer.
B3GNT3 expression was significantly increased in the serum of patients with lung adenocarcinoma
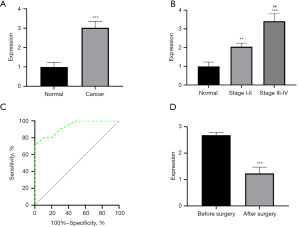
According to the inclusion and exclusion criteria, this study ultimately included serum samples from 30 clinical patients with lung adenocarcinoma and 30 healthy volunteers subjected to propensity matching. Detection of serum B3GNT3 expression by ELISA showed significantly higher B3GNT3 expression levels in patients with lung adenocarcinoma than in normal controls (Figure 2A, P<0.05). The B3GNT3 expression levels of patients with lung adenocarcinoma were compared according to the clinical stage of lung adenocarcinoma, which showed that the expression level of B3GNT3 in tumor tissues of patients with stage I and IIa lung adenocarcinoma was significantly higher than that of normal controls, and the difference was significant. In addition, the comparison of patients with different clinical stages showed that the higher the clinical stage was, the higher the expression level of B3GNT3, and the difference was statistically significant (Figure 2B, P<0.05). The K-M survival curve was drawn according to the expression level of B3GNT3 in lung adenocarcinoma tissue and showed that the prognosis of patients with high B3GNT3 expression was significantly worse than that of patients with low B3GNT3 expression (P<0.05). The areas under the receiver operating characteristic (ROC) curves (AUC) were 0.9373 and 0.9938, which indicated that B3GNT3 had high sensitivity and specificity for the diagnosis of lung adenocarcinoma (Figure 2C). Peripheral serum samples were collected from the patients 1 week before and after the surgery, and B3GNT3 expression in the serum was determined by ELISA. The results showed that B3GNT3 expression significantly decreased in the peripheral serum of patients after surgical resection of tumor tissues compared with that before surgery (Figure 2D, P<0.05). All the above results indicate that the expression level of B3GNT3 is significantly upregulated in the early stage of lung adenocarcinoma. With the progression of clinical stage, the expression of B3GNT3 is further increased, and the expression level of B3GNT3 is closely related to the prognosis of patients. Therefore, B3GNT3 can serve as a potential hematologic marker for lung adenocarcinoma.
B3GNT3 gene silencing suppressed the proliferation of lung adenocarcinoma cells
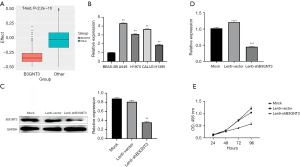
Based on the Cancer Dependency Map (DEPMAP) database, the effects of B3GNT3 and 2,000 random genes on lung adenocarcinoma cells after CRISPR knockdown were compared. The results suggested that B3GNT3 knockout affects lung adenocarcinoma cells more significantly than 2,000 random genes (Figure 3A, P<0.05). Further detection of B3GNT3 expression in different lung adenocarcinoma cells by qPCR showed that B3GNT3 expression in lung adenocarcinoma cells was significantly increased compared with that in normal lung epithelial cells (BEAS-2B cells) (Figure 3B, P<0.05). To explore the effect of B3GNT3 on the proliferative capacity of lung adenocarcinoma cells, the representative cell line A549 was selected from lung adenocarcinoma cells to construct the A549 cell line by constructing adenovirus transfection, and the efficiency of B3GNT3 knockdown was verified by qPCR and WB (Figure 3C, P<0.05). The medium was collected from the culture of knockdown cells and control cells, and the secreted B3GNT3 protein in the medium was detected with ELISA, showing that compared with the controls, B3GNT3 knockout cells secreted significantly less B3GNT3 (Figure 3D, P<0.05). Cell proliferation was detected by CCK-8 assay with the B3GNT3 knockdown cell line, which showed that compared with that of the control cells, the proliferation capacity of B3GNT3 knockout cells was significantly reduced (Figure 3E, P<0.05). These results suggest that suppression of B3GNT3 expression can significantly suppress the proliferation of lung adenocarcinoma cells.
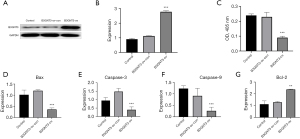
The apoptosis level increased in A549 cells after knocking down B3GNT3
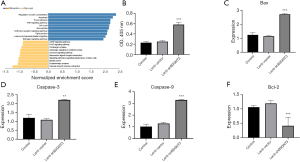
Lung adenocarcinoma samples were divided into high and low B3GNT3 expression groups according to the median B3GNT3 expression, and the differentially expressed genes between the high and low B3GNT3 expression groups were obtained. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment pathway analysis was conducted according to the acquired differentially expressed genes, which showed that the differentially expressed genes were mainly enriched in apoptosis-associated pathways (Figure 4A). The A549 cell line was cultured in vitro, and B3GNT3 expression in A549 cells was knocked down by adenovirus. The results of the apoptosis measured by ELISA showed that the expression of B3GNT3 was significantly increased after overexpression of B3GNT3; the expression of B3GNT3 was also significantly increased after overexpression of B3GNT3 and inhibition of PD-L1 (Figure 4B, P<0.05). RT-PCR showed that the expression of apoptosis-associated genes, including Bax (Figure 4C), caspase 3 (Figure 4D) and caspase 9 (Figure 4E), significantly increased (P<0.05), and the expression of the antiapoptotic gene Bcl-2 significantly decreased (Figure 4F, P<0.05) in A546 cells. The results of the above experiments suggest that B3GNT3 can suppress the apoptosis of lung adenocarcinoma cells.
The apoptosis level of A549 cells was decreased after overexpressing B3GNT3
To further clarify the effect of B3GNT3 on the apoptosis of A549 cells, a B3GNT3 mimic (overexpression) lentivirus was constructed to infect the A549 cell line. B3GNT3 expression in cells was measured by WB (Figure 5A), and the results showed a significant increase in B3GNT3 expression in cells after overexpressing B3GNT3 (Figure 5B, P<0.05). The apoptosis of cells in each group detected by ELISA showed a significant decrease in the apoptosis rate after overexpressing B3GNT3 (Figure 5C, P<0.05). RT-PCR showed that the expression levels of apoptosis-associated genes, including Bax (Figure 5D), caspase 3 (Figure 5E) and caspase 9 (Figure 5F), decreased significantly (P<0.05), while the expression of the proliferation-associated gene Bcl-2 increased significantly (Figure 5G, P<0.05). After overexpressing B3GNT3 in A549 cells, the proliferation capacity of the cells increased significantly, while the apoptosis level was significantly decreased.
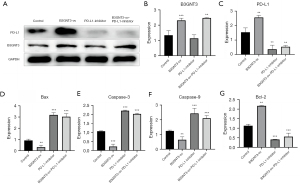
B3GNT3 regulates the apoptosis of lung adenocarcinoma cells via PD-L1
To further clarify the specific molecular mechanism by which B3GNT3 regulates the apoptosis of lung adenocarcinoma cells, a B3GNT3 mimic lentivirus was constructed to overexpress B3GNT3 in A549 cells. PD-L1 expression in cells was suppressed by adding a PD-L1 inhibitor, and the expression of B3GNT3 and PD-L1 in each group was detected by WB at the same time (Figure 6A). The results showed a significant increase in B3GNT3 expression after PD-L1 inhibition in cells with overexpression of B3GNT3 (Figure 6B, P<0.05). PD-L1 expression in cells significantly increased after overexpressing B3GNT3 and was significantly suppressed after inhibiting PD-L1 or both overexpressing B3GNT3 and inhibiting PD-L1 (Figure 6C, P<0.05). RT-PCR showed that the expression levels of the apoptosis-associated genes, including Bax (Figure 6D), caspase 3 (Figure 6E) and caspase 9 (Figure 6F), significantly decreased after overexpressing B3GNT3 (P<0.05), and Bcl-2 expression significantly increased (Figure 6G, P<0.05). The expression levels of apoptosis-associated genes, including Bax (Figure 6D), caspase 3 (Figure 6E) and caspase 9 (Figure 6F), increased significantly after inhibiting PD-L1 (P<0.05). Bcl-2 expression significantly decreased (Figure 6G, P<0.05). The expression of apoptosis-associated genes, including Bax (Figure 6D), caspase 3 (Figure 6E) and caspase 9 (Figure 6F), in cells increased significantly after overexpression of B3GNT3 combined with inhibition of PD-L1 (P<0.05). Bcl-2 expression significantly decreased (Figure 6G, P<0.05). The above results indicate that B3GNT3 regulates the apoptosis of lung adenocarcinoma cells via PD-L1.
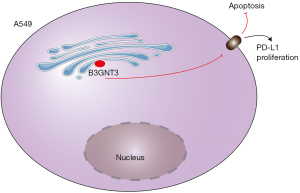
Discussion
In this study, tumor samples from patients with lung adenocarcinoma and normal control samples were obtained from the TCGA database, and the key secreted protein, B3GNT3, which is closely related to the occurrence and development of lung adenocarcinoma, was examined. The close correlation between B3GNT3 and the prognosis of patients with lung adenocarcinoma was further clarified by human serological data detection and in vitro cell experiments. Further mechanistic studies showed that B3GNT3 can regulate the apoptosis of lung adenocarcinoma cells and then regulate the progression of lung adenocarcinoma. The specific molecular regulation mechanism is detailed in Figure 7.
Lung cancer is one of the most common malignant tumors in the world; it also has a higher incidence in China than in other countries, and its mortality rate is the highest among all malignant tumors in China. Its early symptoms are not obvious in patients, and when the symptoms appear, generally, the disease has entered the advanced stage or metastasized, at which point the survival rate is low and the prognosis is poor. Chemotherapy and targeted treatment can significantly improve the prognosis in patients with lung cancer, but patients still inevitably develop drug resistance and relapse (14,15). Therefore, it is important to achieve early diagnosis of lung adenocarcinoma patients and achieve effective prediction of patient prognosis. Many previous studies have tried to screen serum biomarkers of liver cancer, esophageal cancer and gastric cancer from the TCGA database, and the experimental results show that the selected markers have a good predictive ability for tumors (16-18). On this basis, in this study, 535 tumor samples and 59 normal control samples were obtained after searching the TCGA database, and differentially expressed genes were identified after analyzing mRNA data from the two sample groups. Differentially expressed genes were cross-compared with the secreted protein data obtained from the MetaSecKB database, and ultimately, B3GNT3 was identified as the most significantly differentially expressed secreted protein.
B3GNT3 is a type II transmembrane protein located in the Golgi apparatus. It is involved in the biosynthesis of the polyacetamide chain, generation of principal components of dimeric sialic acid Lewis A, regulation of l-selectin ligand function, lymphocyte trafficking and attribution of T cells (19-22). The B3GNT3 gene is located on chromosome 19q13.1 and belongs to the β3GlcNAcT gene family, which contains at least 8 different β3GlcNAcT genes. Previous studies suggest that β3GlcNAcT family members are closely related to the occurrence and development of malignant tumors (23-25). B3GNT3 was found to be closely related to the occurrence and development of breast cancer (26), but there was no report on its relationship with lung adenocarcinoma. In this study, various information of lung adenocarcinoma patients was obtained from the TCGA database at first, and then the patients were divided into groups with high B3GNT3 expression and low B3GNT3 expression. K-M survival curve analysis revealed that the prognosis of patients with high B3GNT3 expression was significantly worse than that of patients with low B3GNT3 expression (P<0.05). The above results indicate that B3GNT3 expression has significant value in predicting the prognosis of patients. The research team further detected the expression of B3GNT3 in serum samples of lung adenocarcinoma patients and healthy volunteers by ELISA. The results showed that the expression level of B3GNT3 in the peripheral blood of lung adenocarcinoma patients was significantly higher than that in normal controls. The ROC curve was drawn, and the AUC values were calculated to be 0.9373 and 0.9938, which proved that the expression of B3GNT3 in serum could be used as a biomarker for the diagnosis of lung adenocarcinoma. Subsequently, a subgroup analysis of lung adenocarcinoma patients was performed according to their clinical stage. The results showed that the serum B3GNT3 level in patients with stage I and IIa lung adenocarcinoma was significantly higher than that in the control group, and with the increase in clinical stage, the expression level of B3GNT3 also increased. Therefore, B3GNT3 can be used as an important serum biomarker for the early screening of patients with lung adenocarcinoma. B3GNT3 can also be used as a serum biomarker for prediction of the prognosis of patients with lung adenocarcinoma.
To further clarify the specific molecular mechanism by which B3GNT3 regulates tumor progression in patients with lung adenocarcinoma, this study divided patients with lung adenocarcinoma into high and low B3GNT3 expression groups according to mRNA information obtained from the TCGA database. KEGG analysis of differentially expressed genes in the two groups showed that the differentially expressed genes were mainly enriched in apoptosis-associated signaling pathways. To further clarify the regulatory effect of B3GNT3 on the apoptosis of lung adenocarcinoma cells, this study attempted to detect changes in proliferation and apoptosis by cultivating the cells in vitro. Moreover, B3GNT3 knockdown and overexpression lentiviruses were constructed and used to transduce lung adenocarcinoma cells, and changes in the proliferation and apoptosis of cells were detected. The results showed that the apoptosis rate of cells increased significantly after knocking down B3GNT3 and decreased significantly after overexpressing B3GNT3. Through bioinformatics and in vitro cell assays, this study clarified that B3GNT3 can regulate the disease progression of lung adenocarcinoma by suppressing the apoptosis of lung adenocarcinoma cells.
To further clarify the specific molecular mechanism of B3GNT3 in lung adenocarcinoma cells, this study attempted to use a PD-L1 inhibitor to overexpress B3GNT3. As one of the PD-1 ligands, PD-L1 can be expressed constitutively in different tissues under physiological conditions (27), mainly in activated T lymphocytes, B lymphocytes, dendritic cells (DC), monocytes, mesenchymal stem cells (MSCs), bone marrow (BM)-derived mast cells and various immune privileged organs (28,29). However, in the tumor immune microenvironment, the PD-1/PD-L1 axis is hijacked by cancer cells to evade immune surveillance. PD-L1 is expressed on different types of tumor cells, including glioblastoma, ovarian cancer, renal cell carcinoma, oral squamous cell carcinoma, colon carcinoma, and NSCLC cells (30-32). With the deepening of PD-L1 research, researchers have found that PD-L1 can also effectively suppress the apoptosis of tumor cells, thus promoting disease progression in tumor patients (33). To further clarify the specific molecular mechanism of PD-L1 and B3GNT3 in the apoptosis of lung adenocarcinoma cells, in this study, a PD-L1 inhibitor was added to B3GNT3 overexpression to detect the changes in apoptosis by cell assays. The results showed that apoptosis decreased after overexpressing B3GNT3, increased after inhibiting PD-L1, and increased after overexpressing PD-L1. The above findings suggest that B3GNT3 can regulate the apoptosis of lung adenocarcinoma cells via PD-L1.
Conclusions
B3GNT3 is an effective serum biologic marker for the evaluation of lung adenocarcinoma. Moreover, B3GNT3 regulates the apoptosis of lung adenocarcinoma cells via PD-L1.
Acknowledgments
Funding: This study was funded by the Clinical Research Application Project, Zhejiang Provincial Health and Health Commission (No. 2022KY479) and Heilongjiang Provincial Department of Education Science and Technology Research Project (No. 2016-KYYWF-0887).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1175/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1175/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1175/coif). All authors report that this study was funded by the Clinical Research Application Project, Zhejiang Provincial Health and Health Commission (No. 2022KY479) and Heilongjiang Provincial Department of Education Science and Technology Research Project (No. 2016-KYYWF-0887). The authors have no other conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Rodak O, Peris-Díaz MD, Olbromski M, et al. Current Landscape of Non-Small Cell Lung Cancer: Epidemiology, Histological Classification, Targeted Therapies, and Immunotherapy. Cancers (Basel) 2021;13:4705. [Crossref] [PubMed]
- Gobbini E, Bertolaccini L, Giaj-Levra N, et al. Epidemiology of oligometastatic non-small cell lung cancer: results from a systematic review and pooled analysis. Transl Lung Cancer Res 2021;10:3339-50. [Crossref] [PubMed]
- Takenaka Y, Oya R, Takemoto N, et al. Predictive impact of sarcopenia in solid cancers treated with immune checkpoint inhibitors: a meta-analysis. J Cachexia Sarcopenia Muscle 2021;12:1122-35. [Crossref] [PubMed]
- Stein JN, Rivera MP, Weiner A, et al. Sociodemographic disparities in the management of advanced lung cancer: a narrative review. J Thorac Dis 2021;13:3772-800. [Crossref] [PubMed]
- Göker E, Altwairgi A, Al-Omair A, et al. Multi-disciplinary approach for the management of non-metastatic non-small cell lung cancer in the Middle East and Africa: Expert panel recommendations. Lung Cancer 2021;158:60-73. [Crossref] [PubMed]
- Kanthaje S, Baikunje N, Kandal I, et al. Repertoires of MicroRNA-30 family as gate-keepers in lung cancer. Front Biosci (Schol Ed) 2021;13:141-56. [Crossref] [PubMed]
- Wu HX, Zhuo KQ, Wang K. Efficacy of targeted therapy in patients with HER2-positive non-small cell lung cancer: A systematic review and meta-analysis. Br J Clin Pharmacol 2022;88:2019-34. [Crossref] [PubMed]
- Naim N, Moukheiber S, Daou S, et al. KRAS-G12C covalent inhibitors: A game changer in the scene of cancer therapies. Crit Rev Oncol Hematol 2021;168:103524. [Crossref] [PubMed]
- Zhou Q, Liu L, Zhou J, et al. Novel Insights Into MALAT1 Function as a MicroRNA Sponge in NSCLC. Front Oncol 2021;11:758653. [Crossref] [PubMed]
- Aggarwal C, Bubendorf L, Cooper WA, et al. Molecular testing in stage I-III non-small cell lung cancer: Approaches and challenges. Lung Cancer 2021;162:42-53. [Crossref] [PubMed]
- Jiang M, Jia K, Wang L, et al. Alterations of DNA damage response pathway: Biomarker and therapeutic strategy for cancer immunotherapy. Acta Pharm Sin B 2021;11:2983-94. [Crossref] [PubMed]
- Chu Q, Gu X, Zheng Q, et al. Long noncoding RNA SNHG4: a novel target in human diseases. Cancer Cell Int 2021;21:583. [Crossref] [PubMed]
- Sun D, Teng F, Xing P, et al. ARID1A serves as a receivable biomarker for the resistance to EGFR-TKIs in non-small cell lung cancer. Mol Med 2021;27:138. [Crossref] [PubMed]
- Pacini L, Jenks AD, Lima NC, et al. Targeting the Fibroblast Growth Factor Receptor (FGFR) Family in Lung Cancer. Cells 2021;10:1154. [Crossref] [PubMed]
- Galvano A, Gristina V, Malapelle U, et al. The prognostic impact of tumor mutational burden (TMB) in the first-line management of advanced non-oncogene addicted non-small-cell lung cancer (NSCLC): a systematic review and meta-analysis of randomized controlled trials. ESMO Open 2021;6:100124. [Crossref] [PubMed]
- Indini A, Rijavec E, Grossi F. Circulating Biomarkers of Response and Toxicity of Immunotherapy in Advanced Non-Small Cell Lung Cancer (NSCLC): A Comprehensive Review. Cancers (Basel) 2021;13:1794. [Crossref] [PubMed]
- Wang Y, Wang J, Wang C, et al. DIO3OS as a potential biomarker of papillary thyroid cancer. Pathol Res Pract 2022;229:153695. [Crossref] [PubMed]
- Li H, Zhang Y, Zheng S. Comprehensive Analysis Identified ETV7 as a Potential Prognostic Biomarker in Bladder Cancer. Biomed Res Int 2021;2021:8530186. [Crossref] [PubMed]
- Zhang H, Lu D, Li Q, et al. Identification of Six Prognostic Genes in EGFR-Mutant Lung Adenocarcinoma Using Structure Network Algorithms. Front Genet 2021;12:755245. [Crossref] [PubMed]
- Wang JS, Ruan F, Guo LZ, et al. B3GNT3 acts as a carcinogenic factor in endometrial cancer via facilitating cell growth, invasion and migration through regulating RhoA/RAC1 pathway-associated markers. Genes Genomics 2021;43:447-57. [Crossref] [PubMed]
- Kong K, Zhao Y, Xia L, et al. B3GNT3: A prognostic biomarker associated with immune cell infiltration in pancreatic adenocarcinoma. Oncol Lett 2021;21:159. [Crossref] [PubMed]
- Zhuang H, Zhou Z, Zhang Z, et al. B3GNT3 overexpression promotes tumor progression and inhibits infiltration of CD8+ T cells in pancreatic cancer. Aging (Albany NY) 2020;13:2310-29. [Crossref] [PubMed]
- Li S, Gao J, Hou L, et al. The Small Molecule Fractions of Floccularia luteovirens Induce Apoptosis of NSCLC Cells through Activating Caspase-3 Activity. Int J Mol Sci 2021;22:10609. [Crossref] [PubMed]
- Shi L, Xiong Y, Hu X, et al. BRD4 inhibition promotes TRAIL-induced apoptosis by suppressing the transcriptional activity of NF-κB in NSCLC. Int J Med Sci 2021;18:3090-6. [Crossref] [PubMed]
- Hu H, Zhang XW, Li L, et al. Inhibition of autophagy by YC-1 promotes gefitinib induced apoptosis by targeting FOXO1 in gefitinib-resistant NSCLC cells. Eur J Pharmacol 2021;908:174346. [Crossref] [PubMed]
- Liu S, Polsdofer EV, Zhou L, et al. Upregulation of endogenous TRAIL-elicited apoptosis is essential for metformin-mediated antitumor activity against TNBC and NSCLC. Mol Ther Oncolytics 2021;21:303-14. [Crossref] [PubMed]
- Gounant V, Brosseau S, Zalcman G. Immunotherapy, the promise for present and future of malignant pleural mesothelioma (MPM) treatment. Ther Adv Med Oncol 2021;13:17588359211061956. [Crossref] [PubMed]
- Yu J, Zhang Q, Li J, et al. Sequential administration of pemetrexed and cisplatin reprograms tumor immune microenvironment and potentiates PD-1/PD-L1 treatment in a lung cancer model. J Investig Med 2022;70:792-9. [Crossref] [PubMed]
- Xia Y, Wang WC, Shen WH, et al. Thalidomide suppresses angiogenesis and immune evasion via lncRNA FGD5-AS1/miR-454-3p/ZEB1 axis-mediated VEGFA expression and PD-1/PD-L1 checkpoint in NSCLC. Chem Biol Interact 2021;349:109652. [Crossref] [PubMed]
- Xie C, Zhou X, Liang C, et al. Apatinib triggers autophagic and apoptotic cell death via VEGFR2/STAT3/PD-L1 and ROS/Nrf2/p62 signaling in lung cancer. J Exp Clin Cancer Res 2021;40:266. [Crossref] [PubMed]
- Chen Z, Chen Z, Xu S, et al. LncRNA SOX2-OT/miR-30d-5p/PDK1 Regulates PD-L1 Checkpoint Through the mTOR Signaling Pathway to Promote Non-small Cell Lung Cancer Progression and Immune Escape. Front Genet 2021;12:674856. [Crossref] [PubMed]
- Jiang ZB, Wang WJ, Xu C, et al. Luteolin and its derivative apigenin suppress the inducible PD-L1 expression to improve anti-tumor immunity in KRAS-mutant lung cancer. Cancer Lett 2021;515:36-48. [Crossref] [PubMed]
- Luo Y, Ma S, Sun Y, et al. MUC3A induces PD-L1 and reduces tyrosine kinase inhibitors effects in EGFR-mutant non-small cell lung cancer. Int J Biol Sci 2021;17:1671-81. [Crossref] [PubMed]


