
SNHG 12 and hsa-miR-140-5P may play an important role in the ceRNA network related to hypertrophic cardiomyopathy
Highlight box
Key findings
• SNHG12, hsa-miR-140-5p, hsa-miR-217, TFRC, HDAC4, TJP1, IGFBP5, and CREB5 may form an important network involved in the pathology of HCM.
What is known and what is new?
• HCM is an inherited heart disease, with diverse and complex phenotypic and genetic expression, yet the precise mechanisms of HCM have not been fully analyzed.
• We constructed a potential ceRNA network including 8 lncRNAs (e.g., LINC00324, SNHG12, and ALMS1-IT1), 7 miRNAs (e.g., hsa-miR-217, hsa-miR-184, and hsa-miR-140-5p) and 52 mRNAs (e.g., IGFBP5, TMED5, and MAGT1) related to HCM.
What is the implication, and what should change now?
• We found that downregulation of SNHG12 and the consequent upregulation of hsa-miR-217, hsa-miR-140-5p, and hsa-miR-508-3p and downregulation of hsa-miR-1244 and hsa-miR-876-3p may play important roles in the pathology of HCM. Further study should focus on this network to provide sufficient evidence.
Introduction
Hypertrophic cardiomyopathy (HCM) is an inherited heart disease, with diverse and complex phenotypic and genetic expression, and has a prevalence of 1:200 to 1:500 (1,2). At least 8 genes and more than 1,500 mutations have been identified to influence left ventricular hypertrophy (LVH) (1,3). HCM is one of the leading causes of sudden cardiac death and occurs in asymptomatic patients. Although surgical treatment is possible, there is no perfect drug-based solution. Among patients with HCM and a pathogenic sarcomeric gene variant, 70% are involved in beta myosin heavy chain 7 (MYH7) and myosin-binding protein C3 (MYBPC3), whereas other genes (MYL2, TNNI3, TNNT2, MYL3, ACTC1, TPM1) each account for a small proportion of patients (1–5%) (4,5). The precise mechanisms of HCM have not been fully analyzed.
Long non-coding RNAs (lncRNAs), microRNAs (miRNAs) and messenger RNAs (mRNAs) form large networks called competing endogenous RNAs (ceRNAs). LncRNAs can act as endogenous molecules and bind small RNA competitively by sharing miRNA response elements with reverse complementary binding seed regions to indirectly regulate mRNA expression levels (6). Recently, an increasing number of studies on ceRNAs have yielded new concepts of gene-related diseases. Some recent research has shown susceptibility modules and genes in HCM, but the whole ceRNA network has not been established (7).
In this study, we first compared differentially expressed mRNAs (DEmRNAs), miRNAs (DEMs), and lncRNAs (DELs) between HCM tissues and normal tissues. The differentially expressed genes (DEGs) intersecting with 163 miRNAs, 93 lncRNAs, and 432 mRNAs constructed a ceRNA regulatory network of HCM. Then, we used the miRcode, TargetScan, miRTarBase, and miRDB databases to determine the interactions among all the lncRNAs. Finally, we identified 8 lncRNAs, 7 miRNAs, and 52 mRNAs to build the ceRNA network associated with HCM. The workflow of this study is shown in Figure 1. This ceRNA network may lay a useful foundation for further research to reveal the full mechanism of HCM. We present the following article in accordance with the STREGA reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-189/rc).
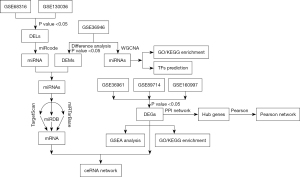
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Data downloading
First, we searched the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geoprofiles) for datasets related to HCM by using the keyword “hypertrophic cardiomyopathy”. The datasets GSE68316, GSE130036, GSE36946, GSE36961, GSE89714, and GSE160997 were downloaded. LncRNA expression profiling related to HCM was conducted for 7 HCM patients and 5 disease-free individuals who were included in GSE68316 (platform: GPL20113 Capital Bio Human LncRNA Microarray v2.0; CapitalBio, Beijing, China) and for 28 HCM patients and 9 healthy donors who were included in GSE130036 (platform: GPL20795, HiSeq X Ten; Illumina, San Diego, CA, USA). MiRNA expression profiling related to HCM was conducted for 107 HCM patients and 20 control donors who were enrolled in GSE36946 (platform: GPL8179 Illumina Human v2 MicroRNA expression bead chip; Illumina). Gene/mRNA expression profiling in HCM was conducted for 106 cardiac tissues from patients with HCM and 39 control donor cardiac tissues in GSE36961 (platform: GPL15389 Illumina HumanHT-12 V3.0 expression bead chip; Illumina) and for samples from 5 HCM patients and 4 disease-free individuals in GSE89714 (platform: GPL11154 Illumina HiSeq 2000; Illumina) and 18 HCM patients and 5 healthy controls in GSE160997 (platform: GPL11154 Illumina HiSeq 2000).
Statistical analysis
Differential expression screening
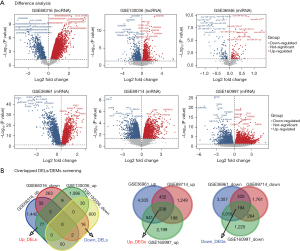
First, the DELs (P<0.05) in myocardial tissues between HCM patients and disease-free individuals were screened for GSE68316 (screened with GEO2R) and GSE130036 (screened with the limma R package; R Foundation for Statistical Computing, Vienna, Austria), and the overlapping lncRNAs in GSE120584 and GSE147232 were selected as the DELs for further study. At the same time, the DEMs (GSE36946) in HCM and normal tissues were screened with GEO2R. The DEMs between HCM and normal tissues were also screened with GEO2R (GSE36961), the limma R package (GSE89714), and the DEseq2 R package (GSE160997). A P value <0.05 was considered to indicate statistical significance. The overlapping mRNAs among GSE36961, GSE89714, and GSE160997 were selected as DEGs.
Weighted gene coexpression network analysis (WGCNA) and functional enrichment analysis of miRNAs
The GSE36946 dataset was downloaded, and WGCNA of DEMs was performed with the R package “WGCNA”. We chose 7 as the soft-thresholding power and 0.8 as the correlation coefficient threshold. Then, we performed functional enrichment analysis [Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG)] and transcription factor (TF) prediction. Herein, P<0.05 was considered to indicate statistical significance.
Functional enrichment analysis of DEGs
To explore the possible biological processes (BPs) involved in HCM, gene set enrichment analysis (GSEA) was performed with clusterProfiler R packages. Padj<0.05 was selected as the threshold. In addition, to forecast the possible functions of the DEGs, KEGG pathway, and GO analyses of the DEGs were conducted with clusterProfiler R packages. GO analysis was conducted for 3 different categories: the BP, cellular component (CC), and molecular function (MF) categories. A P value <0.05 was considered to indicate statistical significance.
Hub gene screening and Pearson correlation network construction
A protein-protein interaction (PPI) network was established using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (https://www.string-db.org/). PPI node pairs with a combined score ≥0.7 were selected for further analysis. Then, the hub genes in the PPI network were screened with CytoHubba software (https://apps.cytoscape.org/apps/cytohubba). The hub genes were visualized with Cytoscape software (https://cytoscape.org/). In addition, the hub genes were used to construct the Pearson correlation network.
ceRNA network construction
Herein, we used the ggalluvial R package to construct lncRNA–miRNA–mRNA triads through miRcode, miRDB, miRTarBase, and TargetScan on the basis of the DEGs and DELs.
First, we predicted the lncRNA-miRNA pairs through the miRcode database on the basis of DELs. Then, we used the miRDB, miRTarBase, and TargetScan databases to assess the characteristics of the miRNA target genes. Any 2 genes that existed in the database were considered miRNA target genes. At last, the (lncRNA–miRNA–mRNA) ceRNA network is constructed using only the remaining overlapping genes and their interacting pairs by comparing the predicted target genes with essential genes composed of DEGs.
GO and KEGG enrichment analysis of the ceRNA network
To forecast the possible functions of this ceRNA network, KEGG pathway and GO analyses were conducted with the clusterProfiler R package. A P value <0.05 was considered statistically significant.
Results
Differential expression screening
As shown in Figure 2A,2B, 93 DELs (77 up-regulated and 16 down-regulated), 163 DEMs (91 up-regulated and 72 down-regulated), and 432 DEGs were screened. They are used in the subsequent analysis.
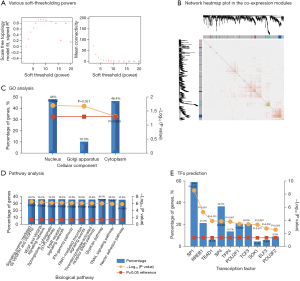
WGCNA and functional enrichment analysis of miRNAs
We used 0.8 as the correlation coefficient threshold, so the soft-thresholding power was 7 (Figure 3A). Through WGCNA (all miRNAs), many coexpression modules were constructed. These modules were independent of other modules (Figure 3B). In this, 4 dominant module genes (yellow, turquoise, brown, and blue modules, including 81 genes) were used for the next analysis. As shown in Figure 3C-3E, the functional enrichment analysis results of miRNAs showed that they were mainly related to the VEGFR signaling network (A), the sphingosine 1-phosphate pathway (B), and the IFNγ pathway (C). These miRNAs were mainly regulated by TFs such as SP1, SP4, RREB1, and POU2F1.
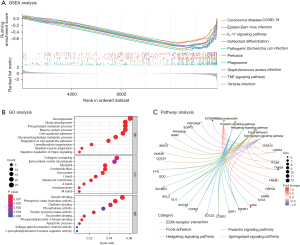
Functional enrichment analysis of DEGs
As shown in Figure 4A, the GSEA results indicated that the DEGs were mainly enriched for the coronavirus disease of 2019 (COVID-19), Epstein-Barr virus infection, the IL-17 signaling pathway, osteoclast differentiation, and TNF signaling pathway terms. As illustrated in Figure 4B,4C, the significantly enriched HCMrelated GO terms mainly included the following: tubulin binding, phosphoric ester hydrolase activity, and cadherin binding in the MF category; axonogenesis, gland development, and phospholipid metabolic process in the BP category; and collagen-containing extracellular matrix (ECM), microtubule, and myofibril in the CC category. KEGG analysis showed that the top 5 pathways related to HCM were the ECM-receptor interaction, focal adhesion, prolactin signaling, Hedgehog signaling, and sphingolipid signaling pathways. Interactions between cells and the ECM are mediated by transmembrane molecules composed primarily of integrins, proteoglycans, CD36, or other cell surface-associated components. These interactions lead to control of cellular activity directly or indirectly, such as adhesion, differentiation, migration, proliferation, and apoptosis. As in HCM, the ECM-receptor interaction is the most significant in the intramuscular fat (8).
Hub gene screening and Pearson correlation network construction
First, 195 nodes and 203 edges were mapped in the PPI networks of DEGs (Figure 5A). Subsequently, the top 21 hub genes of the 3 PPI networks were screened, and these 21 hub genes were used for subsequent Pearson correlation network construction. Finally, as illustrated in Figure 5B, CCND1, LRP5, JAK2, JAK2, and WWC1 [|coefficient| >0.6 and false discovery rate (FDR) <0.05] were screened.
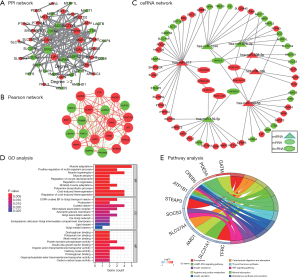
ceRNA network construction
As illustrated in Figure 5C, a ceRNA network including 8 lncRNAs [e.g., LINC00324, small nucleolar RNA host gene 12 (SNHG12), and ALMS1-IT1], 7 miRNAs (e.g., has-miR-217, hsa-miR-184, and hsa-miR-140-5p), and 52 mRNAs (e.g., IGFBP5, TMED5, and MAGT1) was constructed. Of these, 6 lncRNAs were upregulated, 3 corresponding miRNAs were downregulated, and 29 corresponding mRNAs were upregulated in HCM, whereas 2 lncRNAs were downregulated, 4 corresponding miRNAs were upregulated, and 23 corresponding mRNAs were downregulated. These results suggest that lncRNAs may regulate the expression of some mRNAs via certain miRNAs in the context of HCM.
Functional enrichment analysis of the ceRNA network
To better understand the ceRNA network, the KEGG database was utilized to perform GO function and KEGG pathway enrichment analyses. The GO functional annotations included 3 categories, namely, the BP, CC, and MF categories. The top 30 enriched GO terms are listed in Figure 5D. GO BP analysis revealed that the ceRNA network was significantly enriched for the muscle adaptation, muscle hypertrophy, and cold-induced thermogenesis terms. CC analysis revealed that the ceRNA network was significantly enriched for the coated vesicle, COPII-coated ER-to-Golgi transport vesicle, and microtubule-associated complex terms. KEGG pathway enrichment analysis was further conducted for this ceRNA network. As listed in Figure 5E, this ceRNA network was significantly enriched for the ferroptosis, the TNF signaling pathway, insulin resistance, and the cGMP-PKG signaling pathway terms.
Discussion
Clinical diagnosis of HCM in adult patients is made on the basis of a maximal end-diastolic wall thickness of ≥15 mm anywhere in the left ventricle in the absence of another cause of hypertrophy (9-11). Increased left ventricular thickness is associated with an increased risk of sudden death but not necessarily an increased risk of progression to heart failure. Identifying disease-associated’ genetic mutations to make early diagnoses before clinical symptoms appear may prevent sudden death. Genetic testing panels have shown vast heterogeneity and diverse molecular pathways, with more than 2,000 identified sarcomere mutations. Some of the mutations are pathogenic, but most of them are benign (3,12). Furthermore, more evidence regarding the genotype–phenotype correlations is needed to enable the development of genetic tests or other laboratory tests for diagnosis and prediction.
In recent years, increasing numbers of ceRNA networks including mRNAs and lncRNAs have been shown to be involved in disease processes. Molecular interactions in ceRNA networks can promote homeostasis, but disruption of this balance can lead to cancer or other diseases. We built a network of lncRNA, miRNA, and mRNA of HCM. WGCNA revealed that 4 modules (yellow, turquoise, brown, and blue, including 81 miRNAs) were negatively associated with the development of HCM. GO and KEGG enrichment analyses showed that these miRNAs were mainly related to the VEGFR signaling network, sphingosine 1-phosphate pathway, and IFN-gamma pathway and were regulated by TFs such as SP1, SP4, RREB 1, and POU2F1. As illustrated in Figure 5C, a ceRNA network including 8 lncRNAs (e.g., LINC00324, SNHG12, and ALMS1-IT1), 7 miRNAs (e.g., hsa-miR-217, hsa-miR-184, and hsa-miR-140-5p), and 52 mRNAs (e.g., IGFBP5, TMED5, and MAGT1) was constructed. Totally, 6 lncRNAs were upregulated, 3 corresponding miRNAs were downregulated, and 29 corresponding mRNAs were upregulated in HCM, whereas 2 lncRNAs were downregulated, 4 corresponding miRNAs were upregulated, and 23 corresponding mRNAs were downregulated.
Some of the mRNAs in this ceRNA network have been indicated to mediate cardiomyocyte hypertrophy. cAMP responsive element binding protein 5 (CREB5) belongs to the cAMP response element (CRE)-binding protein family. El Jamali found that hypoxia/reoxygenation induces cardiomyocyte hypertrophy through CREB activation. This effect involves β2-AR-dependent pathways involved in the control of CREB activity downstream of GSK3β (13). Furthermore, He et al.’s analysis showed that CREB5 is a downstream protein of the PI3K/Akt pathway that participates in vascular smooth muscle cells’ proliferation and migration which may lead to cardiomyocyte hypertrophy (14). In our study, we found that downregulation of hsa-miR-1244 and upregulation of hsa-miR-217 reduced upregulation of CREB5, and both effects were related to downregulation of SNH12. Furthermore, the IGFBP5 gene is considered the clinical hub gene of HCM (7). Insulin-like growth factor binding protein 5 (IGFBP5) is a protein coding gene. By binding to IGF, IGFBP5 plays important roles in cell differentiation and transcription (15). The role of IGFBP5 in HCM has not been previously reported, but its enrichment in the renin angiotensin system is thought to be an important regulatory mechanism leading to HCM (16). As shown in Figure 5, IGFBP5 was upregulated by hsa-miR-14-5p and hsa-miR-139-5p; this effect was mediated by SNHG12 and LINC00324. The PPI network (Figure 5A) of the hub genes showed that TFRC, HDAC4, and TJP1 were the most important components in the ceRNA network. Several studies have shown the mechanisms of these molecules leading to myocardial injury or cardiomyopathy (17-20).
In our study, most of the hub genes were regulated by hsa-miR-140-5P, whereas all of them were related to SNHG12 (lncRNA). Among all 7 miRNAs involved in the ceRNA network, 5 miRNAs were regulated by SNHG12, which means that SNHG12 may play a dominant role in the development of myocardial hypertrophy. SNHG12 produces a long RNA that is overexpressed in tumor cells. This RNA may promote tumorigenesis by acting as a sponge for miRNAs. In addition to cancer, SNHG12 has been shown to be involved in cerebral ischemia-reperfusion (I/R) injury; specifically, its upregulation protects against cerebral I/R injury (21). Recently, Lu et al. pointed out that the lncRNA SNHG12 is effective in protecting against hypoxia/reoxygenation-induced injury in myocardial cells (22). Furthermore, SNHG12 is a kind of snoRNAs, and these snoRNAs were predicted to have roles in post-translational modifications and alternative splicing, processes differentially regulated in HCM (23). Based on our study, downregulation of SNHG12 and the consequent upregulation of hsa-miR-217, hsa-miR-140-5p, and hsa-miR-508-3p and downregulation of hsa-miR-1244 and hsa-miR-876-3p may play important roles in the pathology of HCM. In particular, the SNHG12–hsa-miR-140-5p–TFRC/HDAC4/TJP1/IGFBP5 pathway and the 4 mRNAs have been shown to take part in myocardial injury by different mechanisms.
Our research had some innate limitations, like lack of in vivo and in vitro validation due to the difficulty in obtaining heart tissues. Subsequent experiments, such as quantitative polymerase chain reaction (qPCR) and western blot experiments, should be performed in the future. Our use of multiple databases for the analysis is another limitation. Different treatments were used on these heart tissues, and the batch effects could not be eliminated.
Conclusions
We have established a complete ceRNA network of HCM including 8 lncRNAs, 7 miRNAs, and 52 mRNAs. We identified 6 lncRNAs, 4 miRNAs, and 29 mRNAs that were upregulated in HCM, and a further 2 lncRNAs, 3 miRNAs, and 23 mRNAs that were downregulated. Among them, SNHG12, hsa-miR-140-5p, hsa-miR-217, TFRC, HDAC4, TJP1, IGFBP5, and CREB5 may form an important pathological network. These genes may provide new drug targets for the treatment of hypertrophic cardiomyopathy and provide new directions for further research on HCM.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-189/rc
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-189/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-189/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Maron BJ, Maron MS. Hypertrophic cardiomyopathy. Lancet 2013;381:242-55. [Crossref] [PubMed]
- Semsarian C, Ingles J, Maron MS, et al. New perspectives on the prevalence of hypertrophic cardiomyopathy. J Am Coll Cardiol 2015;65:1249-54. [Crossref] [PubMed]
- Gersh BJ, Maron BJ, Bonow RO, et al. 2011 ACCF/AHA guideline for the diagnosis and treatment of hypertrophic cardiomyopathy: executive summary: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation 2011;124:2761-96. [Crossref] [PubMed]
- Ho CY, Day SM, Ashley EA, et al. Genotype and Lifetime Burden of Disease in Hypertrophic Cardiomyopathy: Insights from the Sarcomeric Human Cardiomyopathy Registry (SHaRe). Circulation 2018;138:1387-98. [Crossref] [PubMed]
- Serra W, Vitetta G, Uliana V, et al. Severe hypertrophic cardiomyopathy in a patient with a homozygous MYH7 gene variant. Heliyon 2022;8:e12373. [Crossref] [PubMed]
- Bai Y, Long J, Liu Z, et al. Comprehensive analysis of a ceRNA network reveals potential prognostic cytoplasmic lncRNAs involved in HCC progression. J Cell Physiol 2019;234:18837-48. [Crossref] [PubMed]
- Sun Y, Xiao Z, Chen Y, et al. Susceptibility Modules and Genes in Hypertrophic Cardiomyopathy by WGCNA and ceRNA Network Analysis. Front Cell Dev Biol 2021;9:822465. [Crossref] [PubMed]
- Lee HJ, Jang M, Kim H, et al. Comparative Transcriptome Analysis of Adipose Tissues Reveals that ECM-Receptor Interaction Is Involved in the Depot-Specific Adipogenesis in Cattle. PLoS One 2013;8:e66267. [Crossref] [PubMed]
- Geske JB, Ommen SR, Gersh BJ. Hypertrophic Cardiomyopathy: Clinical Update. JACC Heart Fail 2018;6:364-75. [Crossref] [PubMed]
- Marian AJ, Braunwald E. Hypertrophic Cardiomyopathy: Genetics, Pathogenesis, Clinical Manifestations, Diagnosis, and Therapy. Circ Res 2017;121:749-70. [Crossref] [PubMed]
- Maron BJ. Clinical Course and Management of Hypertrophic Cardiomyopathy. N Engl J Med 2018;379:655-68. [Crossref] [PubMed]
- Nefedova VV, Kopylova GV, Shchepkin DV, et al. Impact of Troponin in Cardiomyopathy Development Caused by Mutations in Tropomyosin. Int J Mol Sci 2022;23:15723. [Crossref] [PubMed]
- El Jamali A, Freund C, Rechner C, et al. Reoxygenation after severe hypoxia induces cardiomyocyte hypertrophy in vitro: activation of CREB downstream of GSK3beta. FASEB J 2004;18:1096-8. [Crossref] [PubMed]
- He W, Wang Y, Yang R, et al. Molecular Mechanism of Naringenin Against High-Glucose-Induced Vascular Smooth Muscle Cells Proliferation and Migration Based on Network Pharmacology and Transcriptomic Analyses. Front Pharmacol 2022;13:862709. [Crossref] [PubMed]
- Poreba E, Durzynska J. Nuclear localization and actions of the insulin-like growth factor 1 (IGF-1) system components: Transcriptional regulation and DNA damage response. Mutat Res Rev Mutat Res 2020;784:108307. [Crossref] [PubMed]
- Musumeci B, Tini G, Russo D, et al. Left Ventricular Remodeling in Hypertrophic Cardiomyopathy: An Overview of Current Knowledge. J Clin Med 2021;10:1547. [Crossref] [PubMed]
- Zhang L, Wang H, Zhao Y, et al. Myocyte-specific overexpressing HDAC4 promotes myocardial ischemia/reperfusion injury. Mol Med 2018;24:37. [Crossref] [PubMed]
- Wang N, Ma H, Li J, et al. HSF1 functions as a key defender against palmitic acid-induced ferroptosis in cardiomyocytes. J Mol Cell Cardiol 2021;150:65-76. [Crossref] [PubMed]
- Jiang H, Jia D, Zhang B, et al. Exercise improves cardiac function and glucose metabolism in mice with experimental myocardial infarction through inhibiting HDAC4 and upregulating GLUT1 expression. Basic Res Cardiol 2020;115:28. [Crossref] [PubMed]
- De Bortoli M, Postma AV, Poloni G, et al. Whole-Exome Sequencing Identifies Pathogenic Variants in TJP1 Gene Associated With Arrhythmogenic Cardiomyopathy. Circ Genom Precis Med 2018;11:e002123. [Crossref] [PubMed]
- Yin WL, Yin WG, Huang BS, et al. LncRNA SNHG12 inhibits miR-199a to upregulate SIRT1 to attenuate cerebral ischemia/reperfusion injury through activating AMPK signaling pathway. Neurosci Lett 2019;690:188-95. [Crossref] [PubMed]
- Lu P, Xiao S, Chen S, et al. LncRNA SNHG12 downregulates RAGE to attenuate hypoxia-reoxygenation-induced apoptosis in H9c2 cells. Biosci Biotechnol Biochem 2021;85:866-73. [Crossref] [PubMed]
- James V, Nizamudeen ZA, Lea D, et al. Transcriptomic Analysis of Cardiomyocyte Extracellular Vesicles in Hypertrophic Cardiomyopathy Reveals Differential snoRNA Cargo. Stem Cells Dev 2021;30:1215-27. [Crossref] [PubMed]
(English Language Editor: J. Jones)


