
Investigation of the function of the novel tumor marker BEND5 in lung adenocarcinoma based on data mining and in vitro analysis
Highlight box
Key findings
• BEND5 expression is low in LUAD and may be associated with poor prognosis, and BEND5 overexpression inhibits LUAD cells via the PPAR signaling pathway.
What is known and what is new?
• BEND5 belongs to the BEN family of structural domains, whose members can be found in several animal proteins. BEND5 is dysregulated in colorectal and breast cancer and is strongly associated with tumor progression and prognosis;
• According to bioinformatics, BEND5 expression was low in LUAD and was related to a poor prognosis. In vitro experiments showed that BEND5 overexpression suppressed LUAD cells and decreased the expression of cell cycle-related proteins. Furthermore, BEND5 triggered the PPAR signaling pathway, and knocking down PPAR reversed the effects of BEND5 overexpression on LUAD cells.
What are the implications of the study, and what should change now?
• BEND5 inhibits the proliferation of lung adenocarcinoma cells via activating PPAR signaling pathways, and it could be used as a prognostic biomarker for patients with the disease.
Introduction
Lung cancer accounts for approximately 2.2 million new diagnoses and 1.79 million deaths every year, making it the most prevalent malignancy worldwide (1). The incidence and mortality rates of lung cancer are constantly rising (2,3). The most prevalent subtype of lung cancer is lung adenocarcinoma (LUAD), which accounts for about 40% of cases (4). Currently, surgical resection is the mainstay treatment for LUAD. However, at the time of presentation, most cases have already progressed to an advanced stage of the disease and are no longer suitable for surgical resection. Furthermore, even if LUAD is detected at an early stage, recurrence and metastasis are extremely common issues (5-7). Despite the significant achievements that have been made in the treatment of LUAD, the 5-year overall survival (OS) rate is still only 15%, which reflects the poor prognosis of the disease (8,9). Therefore, the identification of specific molecular targets holds immense potential for the diagnosis, treatment, and prognosis of LUAD.
BEN domain-containing protein (BEND) encodes a domain protein that plays a pivotal role in the maintenance of DNA chromatin structure. The BEND family has nine members: BEND2, BEND3, BEND4, BEND5, BEND6, BEND7, BEND3P1, BEND3P2, and BEND3P3. BEND is critically important for mediating protein-DNA and protein-protein interactions (PPIs) during chromatin organization and transcription (10,11). BEND5 is located on chromosome 1p33 (12) and encodes a protein comprising 421 amino acid residues. The involvement of BEND proteins in the regulation of gene expression has been observed during early germ cell development, with BEND5 overexpression inducing the differentiation of embryonic stem cells into primordial germ progenitor-like cells in vitro (13). Deregulation of BEND5 has been reported in malignant tumors, including colorectal cancer (14) and breast cancer (15). In colorectal cancer, BEND5 is highly downregulated and exhibits high methylation levels. Apoptosis, autophagy, and differentiation are among the key mechanisms via which BEND5 causes a reduction in cell proportions in colorectal cancer, and its reduced expression is correlated with patients’ clinical status and outcome (14). In breast cancer, Shi et al. found that high expression of BEND5 inhibited tumor cell proliferation and metastasis by interacting with the N-terminal domain (NTD) of RBPJ and further inhibiting Notch signaling (15). The same study also demonstrated that low expression of the BEND5 gene could predict an advanced clinical stage and poor OS outcome in patients with breast cancer (15). However, to the best of our knowledge, no report has ever thoroughly investigated the role of BEND5 in LUAD in a comprehensive and integrated manner.
Bioinformatics research has grown with the development of next-generation sequencing (16,17). Public databases such as The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) are ideal for accessing transcriptomic information, which promotes effective ways of identifying gene signatures (18-20). Many studies have attempted to build risk models for biological features or prognostic assessment of malignant tumors that might have a clinical influence (21-23). In the current study, we used the TCGA-LUAD database to assess the expression and prognostic value of BEND5 in LUAD and performed gene set enrichment analysis (GSEA), Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis, Gene Ontology (GO) enrichment analysis, and immune infiltration analysis. It is hoped our findings will inform a better understanding of the role played by BEND5 in LUAD.
Methods
BEND5 expression in pan-cancer data
To determine the expression of BEND5 in pan-cancer, RNA-seq data were downloaded from the Genotype-Tissue Expression (GTEx) cohort and TCGA database (https://portal.gdc.cancer.gov/) in transcript per million (TPM) format using the University of California Santa Cruz (UCSC) Xena Browser (https://xenabrowser.net) (24,25). The Toil method was then applied for uniform data processing. An evaluation and representation of BEND5 mRNA expression levels in various cancers were performed with the ‘ggplot2’ package (version 3.3.3) in R (version 3.6.3). After being normalized to the TPM format, the data from transcript mapping were log2 converted. In total, 10,534 and 15,776 samples were used for paired and unpaired sample analysis, respectively. The expression levels of BEND5 mRNA were compared between healthy control and tumor groups using the Mann-Whitney U test. Paired sample t-tests were conducted to analyze paired sample data that passed the Shapiro-Wilk normality test (P>0.05). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
BEND5 expression in LUAD samples from the TCGA database
For analysis of BEND5 expression in LUAD samples, level 3 high-throughput sequence (HT-seq) data from patients with LUAD were obtained from the TCGA (https://portal.gdc.cancer.gov/) and UCSC Xena Browser (https://xenabrowser.net) (24,25) databases in fragments per kilobase of TPM mapped reads (FPKM)’ format. Samples without clinical data were eliminated. A further analysis was conducted on the TCGA-LUAD dataset, which comprises 594 samples including 535 LUAD tumor and 59 healthy control samples. Log2 transformations were applied following the conversion of FPKM-formatted RNA-sequencing (RNAseq) data to TPM format. The ‘ggplot2’ package (version 3.3.3) in R (version 3.6.3) was used to evaluate and display BEND5 mRNA expression levels in LUAD. Both paired and unpaired samples were analyzed by applying the paired t-test to paired data that passed the Shapiro-Wilk normality test (P>0.05). The Mann-Whitney U test (Wilcoxon rank sum test) was used to examine unpaired data that failed the normality test (P<0.05).
Analysis of the diagnostic utility of BEND5 mRNA expression in LUAD
The ‘pROC’ (version 1.17.0.1) and ‘ggplot2’ (version 3.3.3) packages in R (version 3.6.3) were applied to examine BEND5 gene expression data using the receiver-operating characteristic (ROC) curve. Clinical status (LUAD tumor versus normal) was selected as an outcome parameter. The X-axis represented the false positive rate (FPR), and the Y-axis represented the true positive rate (TPR). In general, diagnostic tests with an area under the ROC curve (AUC) >0.9 are considered highly accurate; those with an AUC 0.7–0.9 are considered moderately accurate; those with an AUC 0.5–0.7 are considered to have low accuracy; and those with an AUC <0.5 are considered random.
Tumor clinical characteristics of TCGA-LUAD samples
The above-described TCGA-LUAD data served as the foundation for the subsequent analysis. Clinicopathological information, BEND5 mRNA expression data, and general patient characteristics were extracted from the dataset. Based on BEND5 expression level, the LUAD samples were classified into a low BEND5 gene expression group and a high BEND5 gene expression group. The following categorical variables of cases were examined: T stage, N stage, M stage, pathologic stage, primary therapy outcome, age, sex, race, smoker (yes or no), anatomic neoplasm subdivision (left or right sites and peripheral or central lung), OS events, disease-specific survival (DSS) events, and progression-free interval (PFI) events. The clinical characteristics of cases in the low and high BEND5 gene expression groups from the TGCA-LUAD dataset are shown in Table 1.
Table 1
| Characteristic | Low BEND5 expression | High BEND5 expression | P |
|---|---|---|---|
| N | 267 | 268 | |
| T stage, n (%) | <0.001 | ||
| T1 (175 cases) | 67 (12.6) | 108 (20.3) | |
| T2 (289 cases) | 154 (28.9) | 135 (25.4) | |
| T3 (49 cases) | 33 (6.2) | 16 (3.0) | |
| T4 (19 cases) | 12 (2.3) | 7 (1.3) | |
| N stage, n (%) | 0.205 | ||
| N0 (348 cases) | 168 (32.4) | 180 (34.7) | |
| N1 (95 cases) | 50 (9.6) | 45 (8.7) | |
| N2 (74 cases) | 45 (8.7) | 29 (5.6) | |
| N3 (2 cases) | 1 (0.2) | 1 (0.2) | |
| M stage, n (%) | 0.884 | ||
| M0 (361 cases) | 189 (49.0) | 172 (44.6) | |
| M1 (25 cases) | 14 (3.6) | 11 (2.8) | |
| Pathologic stage, n (%) | 0.013 | ||
| Stage I (294 cases) | 129 (24.5) | 165 (31.3) | |
| Stage II (123 cases) | 68 (12.9) | 55 (10.4) | |
| Stage III (84 cases) | 52 (9.9) | 32 (6.1) | |
| Stage IV (26 cases) | 14 (2.7) | 12 (2.3) | |
| Primary therapy outcome, n (%) | 0.031 | ||
| PD (71 cases) | 45 (10.1) | 26 (5.8) | |
| SD (37 cases) | 20 (4.5) | 17 (3.8) | |
| PR (6 cases) | 3 (0.7) | 3 (0.7) | |
| CR (332 cases) | 149 (33.4) | 183 (41.0) | |
| Sex, n (%) | <0.001 | ||
| Female (286 cases) | 123 (23) | 163 (30.5) | |
| Male (249 cases) | 144 (26.9) | 105 (19.6) | |
| Race, n (%) | 0.057 | ||
| Asian (7 cases) | 6 (1.3) | 1 (0.2) | |
| Black or African American (55 cases) | 22 (4.7) | 33 (7.1) | |
| White (406 cases) | 199 (42.5) | 207 (44.2) | |
| Age, n (%) | 0.660 | ||
| ≤65 years (255 cases) | 130 (25.2) | 125 (24.2) | |
| >65 years (261 cases) | 127 (24.6) | 134 (26.0) | |
| Residual tumor, n (%) | 0.777 | ||
| R0 (355 cases) | 173 (46.5) | 182 (48.9) | |
| R1 (13 cases) | 8 (2.2) | 5 (1.3) | |
| R2 (4 cases) | 2 (0.5) | 2 (0.5) | |
| Anatomic neoplasm subdivision, n (%) | 1.000 | ||
| Left (205 cases) | 103 (19.8) | 102 (19.6) | |
| Right (315 cases) | 157 (30.2) | 158 (30.4) | |
| Anatomic neoplasm subdivision 2, n (%) | 0.765 | ||
| Central lung (62 cases) | 32 (16.9) | 30 (15.9) | |
| Peripheral lung (127 cases) | 70 (37.0) | 57 (30.2) | |
| No. of pack years smoked, n (%) | 0.074 | ||
| <40 years (188 cases) | 116 (31.4) | 72 (19.5) | |
| ≥40 years (181 cases) | 94 (25.5) | 87 (23.6) | |
| Smoker, n (%) | <0.001 | ||
| No (75 cases) | 18 (3.5) | 57 (10.9) | |
| Yes (446 cases) | 244 (46.8) | 202 (38.8) | |
| OS event, n (%) | 0.022 | ||
| Alive (343 cases) | 158 (29.5) | 185 (34.6) | |
| Dead (192 cases) | 109 (20.4) | 83 (15.5) | |
| DSS event, n (%) | 0.007 | ||
| Alive (379 cases) | 172 (34.5) | 207 (41.5) | |
| Dead (120 cases) | 72 (14.4) | 48 (9.6) | |
| PFI event, n (%) | 0.240 | ||
| Alive (309 cases) | 147 (27.5) | 162 (30.3) | |
| Dead (226 cases) | 120 (22.4) | 106 (19.8) | |
| Age, median (years) [IQR] | 65 [57, 72] | 66 [60, 72] | 0.299 |
BEND5, BEN domain-containing protein 5; PR, partial response; CR, complete response; PD, progressive disease; SD, stable disease; OS, overall survival; DSS, disease-specific survival; PFI, progression-free interval; IQR, interquartile range.
When the theoretical frequency of a categorical variable was greater than 5 and the sample size exceeded 40, the chi-squared test was employed, while Fisher’s exact test was conducted to determine whether any differences were present between the groups with high and low LUAD expression. When a specific categorical variable did not have a normal distribution (P<0.05), the Wilcoxon rank-sum test was applied. The ‘ggplot2’ package (version 3.3.3) in R (version 3.6.3) was used for statistical analysis.
Correlation analysis of BEND5 expression level and tumor clinical features
Multiple categories of clinical features were examined to analyze their relationships with BEND5 mRNA expression level. If data were normally distributed, the ‘ggplot2’ package (version 3.3.3) in R (version 3.6.3) was applied to perform a one-way analysis of variance (ANOVA; P>0.05, Shapiro-Wilk normality test); otherwise, the Kruskal-Wallis test was performed. Data from 329 LUAD tumor samples were examined. A binary logistic model was also applied to study the relationships between BEND5 expression and tumor clinical features.
Survival analysis
Gene expression profiling interactive analysis (GEPIA; http://gepia.cancer-pku.cn/index.html) was used to plot survival maps of BEND5 expression using pan-cancer data. The Kaplan-Meier (KM) plotter (https://kmplot.com/analysis/) was used to evaluate the relationships of BEND5 gene expression with OS and relapse-free survival in patients with LUAD.
Cox regression with univariate and multivariate survival analysis
To examine the relationship between tumor features and patient prognosis in LUAD, univariate and multivariate Cox regression models were used. To determine OS outcomes, the Cox regression module and the coxph function of the survival package (version 3.2-10) in R (version 3.6.3) were employed. In addition to the categorical variables mentioned above, BEND5 gene expression was included as a tumor characteristic feature.
Forest plots
Based on the results of univariate and multivariate Cox regression analyses [hazard ratio (HR), 95% confidence interval (CI), and P value], two forest plots were created using the ‘ggplot2’ package (version 3.3.3) in R (version 3.6.3). By comparing one level of a binary characteristic with a reference level, the HR reflects the relative mortality risk. In general, an HR value >1 denotes a higher chance of death, whereas an HR value <1 indicates a lower risk.
Nomogram plotting
A LUAD prognosis-related nomogram incorporating clinical characteristics and gene expression was created using the TCGA-LUAD dataset, as well as a predictive nomogram that combined BEND5 expression levels with clinical variables was also built. In the R program (version 3.6.3), the rms (version 6.2-0) and survival (version 3.2-10) packages were used to create the nomogram, which was then analyzed statistically by Cox regression. The nomograms were created to predict the OS of patients with LUAD.
Calibration plot
The survival (version 3.2-10) and rms (version 6.2-0) packages in R were used to create calibration plots. To assess the performance of the nomogram model in terms of accuracy for predicting prognostic outcomes of LUAD, the fit between the actual fraction survival probability and the nomogram-predicted survival probability was calculated at three time points (1, 3, and 5 years). The nomogram would be considered to show perfect prediction accuracy if the solid lines for the 1-, 3- and 5-year time points fitted well at a 45-degree ideal diagonal line.
Identification of genes associated with BEND5 in LUAD
BEND5 gene correlation analysis was carried out using the ‘stat’ package (version 3.6.3) in R (version 3.6.3). Solely using protein-coding genes, the Pearson correlation test was applied to determine if the high and low expression groups were linearly related. The Pearson correlation coefficient (Pearson’s r, also known as the cor_Pearson value) was evaluated, with a cor_Pearso value of 0.90–1.00, 0.70–0.89, 0.40–0.69, 0.10–0.39, and 0.00–0.10 indicating a very strong, strong, moderate, weak, and negligible correlation, respectively. BEND5-correlated genes with P value <0.05 were regarded as significantly correlated genes.
Heat map of the top 20 BEND5-correlated genes
Following their identification, genes that were significantly correlated with BEND5 in LUAD, were listed in descending order of cor_Pearson value, and the first ten were considered to be the top 10 positively correlated genes. The significantly correlated genes were then listed in ascending order of cor_Pearson value, and the first ten were considered to be the top 10 negatively correlated genes. The expression of these 20 associated genes in LUAD samples was visualized as a heat map using the ‘ggplot2’ package (version 3.3.3) in R (version 3.6.3).
GSEA
The genes possessing an absolute correlation coefficient >0.1 and a P value <0.05 were selected for GSEA analysis. Significant genes between LUAD and healthy control samples in the TCGA dataset were identified using ‘DESeq2’ (version 1.26.0) in R (version 3.6.3). To perform GSEA, the log2[fold change (FC)] values were acquired for genes that were significantly correlated with BEND5. The GSEA was performed with the clusterProfiler package (version 3.14.3) in R (version 3.6.3). Further, to find the pathways, the Reactome (REAC) Pathway database, the KEGG pathway database and WikiPathways (WP) database were used. The referenced gene set is located in the MSigDB Collections (https://www.gsea-msigdb.org/gsea/msigdb/collections.jsp#C2) as c2.cp.v7.2.symbols.gmt (Curated). Functional terms were deemed to be significantly enriched on meeting the conditions of P.adjust <0.05, false discovery rate (FDR) or q value <0.25, and |normalized enrichment score (NES)| >1.
Functional enrichment analysis of the top 10 genes significantly associated with BEND5
Significantly correlated genes with the threshold values (|r| >0.4 and P<0.05) were selected for functional enrichment analysis. To identify highly enriched functional terms for BEND5-correlated genes, a functional enrichment analysis was conducted on the top 10 positively and negatively correlated genes. The Benjamini and Hochberg correction was used to calculate adjusted P values for Homo sapiens. GO terms—including biological processes (BP), cellular components (CC), and molecular functions (MF)—and KEGG pathways were considered to be significantly enriched for linked genes on meeting the thresholds of P.adjust <0.05 and q value <0.2. Bubble charts to show the outcomes of the enrichment analyses were created using the ‘ggplot2’ package (version 3.3.3) in R (version 3.6.3).
Cell lines and culture
Five LUAD cell lines (H1299, H460, PC-9, A549, and H1975) and a non-neoplastic lung epithelial cell line (HBE) were obtained from the American Type Culture Collection (ATCC). The H1299 cell line (ATCC number: CRL-5803) is an epithelial-like cell line established from a lymph node metastasis from the lung of a patient who had received prior radiation therapy (URL: https://www.atcc.org/products/crl-5803). The H460 cell line (ATCC number: HTB-177) was isolated from the pleural fluid of a male patient with large cell lung cancer in 1982 (URL: https://www.atcc.org/products/htb-177). The PC-9 (formerly known as PC-14) cell line was derived from a human adenocarcinoma from lung tissue which remains undifferentiated. The A549 cell line (ATCC number: CRM-CCL-185) was initiated through explant culture of lung carcinomatous tissue from a 58-year-old Caucasian male. The H1975 cell line (ATCC number: CRL-5908) is a cell line exhibiting epithelial morphology that was isolated from the lungs of a nonsmoking female with non-small cell lung cancer. The cell lines were grown in RPMI-1640 media (Thermo Fisher Scientific, Waltham, MA, USA) with 10% fetal bovine serum (Gibco, USA) and incubated at 37 ℃ with 5% CO2.
Plasmid transfection experiments
Plasmids were purchased from WZ Biosciences (Shandong, China). Following instructions supplied by the manufacturer, the BEND5 plasmid was transfected utilizing Lipofectamine 3000 (Thermo Fisher Scientific, Inc.). Overexpression of BEND5 was stimulated in LUAD cells to investigate its effects.
Western blot analysis
Western blot analysis was carried out following a previously published protocol (https://www.abcam.cn/protocols/general-western-blot-protocol#8). Quantification of protein concentration was performed by employing the BCA protein assay method (Beyotime, China) after the protein had been extracted using radioimmunoprecipitation assay (RIPA) lysis buffer (Fude Biotechnology, China) supplemented with phosphatase and protease inhibitors (CWBIO, Shanghai, China; Thermo Fisher Scientific). Total protein (30 µg) was separated using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred onto polyvinyl difluoride membranes (Invitrogen, USA), and then blocked with 5% nonfat dry milk in Tris-buffered saline, pH 7.5. Primary antibodies were incubated with the membranes for an extended period. The primary antibodies used were as follows: PPAR (Abcam, UK), BEND5, GAPDH, CDK2 (Proteintech, USA), CDK4, CCND1, and CCNB1 (Cell Signaling Technology, USA). The membranes were immunoblotted at 4 ℃ overnight. A horseradish peroxidase (HRP)-conjugated anti-rabbit or anti-mouse IgG antibody was used as the secondary antibody (Proteintech, USA). Enhanced chemiluminescence reagents (Fude Biotechnology, China) were used to detect the signals.
Cell Counting Kit-8 (CCK-8) and colonogenic assays
The CCK-8 assay was performed as described in a previously reported protocol (https://www.abcam.com/cell-counting-kit-8-wst-8--cck8-ab228554.html). For this assay, cells (103 cells per well) were seeded in 96-well plates. The CCK-8 reagent (Beyotime, China) was added continuously for 5 days at 24-hour intervals. Cell viability was determined by measuring the optical density at a wavelength of 450 nm. For the colonogenic experiment, cells were seeded in 6-well plates with 500 cells/well and cultured for 14 days using 37 ℃ with 5% of CO2. Afterward, the colonies were cleaned three times with phosphate-buffered saline and stained with crystal violet for 15 minutes.
Analysis of correlations between immune cells and BEND5 expression in LUAD
Using Spearman’s statistical technique, the relationship between BEND5 expression and immune cells in LUAD tumor tissues was examined. This analysis was carried out using the GSVA package (version 1.34.0) in R (version 3.6.3). The statistical analysis made use of the single-sample GSEA (ssGSEA) method, which is a built-in technique in the GSVA package. The 24 tumor-infiltrating immune cells (TIICs) under study included B cells, CD8 T cells, cytotoxic cells, dendritic cells (DCs), activated DCs, immature DCs, plasmacytoid DCs, macrophages, mast cells, natural killer (NK) cells, NK CD56bright cells, NK CD56dim cells, neutrophils, T cells, T helper cells, T central memory cells, T effector memory cells, T follicular helper cells, gamma delta T cells, Th1 cells, Th17 cells, Th2 cells, and T regulatory cells. The correlation between BEND5 expression and the 24 TIICs in LUAD samples was visualized using a lollipop plot. Scatter plots were utilized to display the statistically significant relationships between BEND5 expression and specific types of immune cell in LUAD samples.
Analysis of correlations between immune checkpoint genes (ICGs) and BEND5 expression
The 59 ICGs include 23 checkpoint inhibitor genes and 36 checkpoint stimulator genes (26). Using Spearman’s statistical approach, the association between the expression of BEND5 expression and each individual ICG in LUAD was assessed. A correlation coefficient of r>0 and a P value <0.05 indicated a significant positive correlation, while r<0 and P value <0.05 indicated a significant negative correlation. The expression patterns of the 59 ICGs in LUAD tumor samples were visualized as a heat map with the ‘ggplot2’ package (version 3.3.3) in R (version 3.6.3). Scatter plots were also created to display the correlations between BEND5 and ICGs that were confirmed to be statistically significant, again using ‘ggplot2’.
Statistical analysis
GraphPad Prism (version 8.0) was used for data processing. Unless otherwise stated, the data values are presented as the mean with standard deviation (SD). Student’s t-test (two-tailed) was employed for comparing independent survey contrasts between two groups when both SDs were the same, and Student’s t-test with Welch’s correlation was used when the SDs of two groups differed. An ANOVA (one way) was applied when variances between multigroup sample statistics were equal, and Welch’s ANOVA was adopted if the variances were not equal. Statistically significant differences were indicated by P<0.05, P<0.01, or P<0.001.
Results
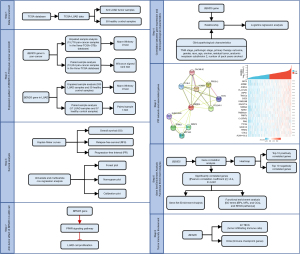
Flowchart of the current study
Figure 1 is a flowchart that depicts the current research study design.
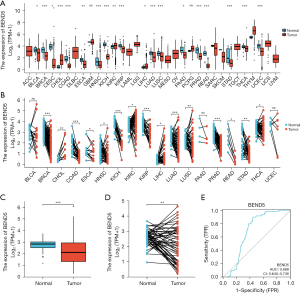
BEND5 dysregulation in pan-cancer data and LUAD
Analysis of pan-cancer data from the TCGA database revealed significant downregulation of BEND5 in multiple cancer types (Figure 2A,2B). Unpaired sample analyses showed that BEND5 was dysregulated in multiple cancers, including breast invasive carcinoma (BRCA), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), kidney chromophobe (KICH), LUAD and pancreatic adenocarcinoma (PAAD), among others (Figure 2A). Paired sample analyses showed that BEND5 was dysregulated in several cancers including BRCA, KICH, kidney renal papillary cell carcinoma (KIRP), LUAD, rectum adenocarcinoma (READ) and stomach adenocarcinoma (STAD), among others (Figure 2B). Further, in comparison with healthy control samples, LUAD tumor samples showed a significant decrease in BEND5 expression (Figure 2C,2D). The ROC curve analysis also showed that BEND5 expression was useful for differentiating LUAD from healthy control samples (AUC =0.688 >0.5; Figure 2E).
Prognostic significance of BEND5 in pan-cancer and LUAD
Data from TCGA were used to determine the prognostic value of BEND5 expression in pan-cancer (Figure 3A,3B). Analysis of the association between BEND5 expression and OS using pan-cancer data showed that elevated BEND5 mRNA expression levels were linked to a poor prognosis in adrenocortical carcinoma (ACC), KICH, READ and STAD but with a good prognosis in BLCA, BRCA, CESC, lymphoid neoplasm diffuse large B-cell lymphoma (DLBC), KIRP, LUAD, PAAD, PRAD, and uveal melanoma (UVM). In terms of relapse-free survival, BEND5 overexpression was found to be linked to a short recurrence time in ACC, KICH, READ, and STAD but to a long recurrence time in BLCA, mesothelioma, PAAD, and UVM. The prognostic value of BEND5 for LUAD was confirmed via the KM method, and the outcome was consistent with earlier findings that BEND5 expression was significantly related to OS and DSS (Figure 3C,3D). Cox regression analysis (univariate and multivariate) was applied to determine whether BEND5 could serve as a biomarker for predicting the prognosis of patients with LUAD independently, and the results revealed that BEND5 expression could strongly predict OS and relapse-free survival in LUAD (Figure 3E,3F, Table 2).
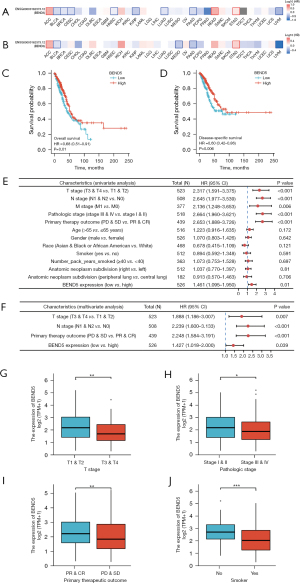
Table 2
| Characteristics | Total (N) | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |||
| T stage (T3 + T4 vs. T1 + T2) | 523 | 2.317 (1.591–3.375) | <0.001 | 1.888 (1.186–3.007) | 0.007 | |
| N stage (N1 + T2 vs. N0) | 508 | 2.645 (1.977–3.539) | <0.001 | 2.239 (1.600–3.133) | <0.001 | |
| M stage (M1 vs. M0) | 377 | 2.136 (1.248–3.653) | 0.006 | – | – | |
| Pathologic stage (stage III + IV vs. stage I + II) | 518 | 2.664 (1.960–3.621) | <0.001 | – | – | |
| Primary therapy outcome (PD + SD vs. PR + CR) | 439 | 2.653 (1.888–3.726) | <0.001 | 2.248 (1.584–3.191) | <0.001 | |
| Age (>65 vs. ≤65 years) | 516 | 1.223 (0.916–1.635) | 0.172 | – | – | |
| Sex (male vs. female) | 526 | 1.070 (0.803–1.426) | 0.642 | – | – | |
| Race (Asian + Black or African American vs. White) | 468 | 0.678 (0.415–1.109) | 0.121 | – | – | |
| Smoker (yes vs. no) | 512 | 0.894 (0.592–1.348) | 0.591 | – | – | |
| No. of pack years smoked (≥40 vs. <40 years) | 363 | 1.073 (0.753–1.528) | 0.697 | – | – | |
| Atomic neoplasm subdivision (right vs. left) | 512 | 1.037 (0.770–1.397) | 0.810 | – | – | |
| Anatomic neoplasm subdivision 2 (peripheral lung vs. central lung) | 182 | 0.913 (0.570–1.463) | 0.706 | – | – | |
| BEND5 expression (high vs. low) | 526 | 1.461 (1.095–1.950) | 0.010 | 1.427 (1.018–2.000) | 0.039 | |
BEND5, BEN domain-containing protein 5; LUAD, lung adenocarcinoma; HR, hazard ratio; CI, confidence interval; PR, partial response; CR, complete response; PD, progressive disease; SD, stable disease.
Correlation between BEND5 expression and clinicopathological characteristics of LUAD
To investigate the putative function of BEND5 in the pathogenesis of LUAD, its expression levels and their association with tumor clinicopathological characteristics were examined. A comparison of tumor characteristics between patients with low and high BEND5 gene expression in LUAD is shown in Table 1, and a comparison of clinical features between these two groups is shown in Table 3. The expression level of BEND5 mRNA in LUAD was significantly linked to four tumor characteristics including T stage, pathologic stage, primary therapy outcome, and smoker (Figure 3G-3J); however, no such correlation was found for other variables such as N stage, M stage, age, race, and the number of pack-years smoked.
Table 3
| Characteristics | Total (N) | Odds ratio (95% CI) | P value |
|---|---|---|---|
| T stage (T3 + T4 vs. T1 + T2) | 532 | 0.578 (0.339–0.968) | 0.039 |
| N stage (N1 + N2 + N3 vs. N0) | 519 | 0.773 (0.535–1.116) | 0.170 |
| M stage (M1 vs. M0) | 386 | 0.873 (0.378–1.970) | 0.744 |
| Pathologic stage (stage III + IV vs. stage I + II) | 527 | 0.656 (0.427–1.001) | 0.052 |
| Primary therapy outcome (PD + SD vs. PR + CR) | 446 | 0.526 (0.336–0.816) | 0.004 |
| Sex (male vs. female) | 535 | 0.622 (0.441–0.875) | 0.007 |
| Race (Asian + Black or African American vs. White) | 468 | 1.179 (0.690–2.028) | 0.548 |
| Age (>65 vs. ≤65 years) | 516 | 1.048 (0.742–1.480) | 0.792 |
| Smoker (yes vs. no) | 521 | 0.286 (0.161–0.489) | <0.001 |
| Residual tumor (R1 + R2 vs. R0) | 372 | 0.658 (0.234–1.751) | 0.406 |
| Anatomic neoplasm subdivision2 (peripheral lung vs. central lung) | 189 | 0.716 (0.388–1.315) | 0.282 |
| No. of pack years smoked (≥40 vs. <40 years) | 369 | 1.334 (0.884–2.018) | 0.171 |
BEND5, BEN domain-containing protein 5; PR, partial response; CR, complete response; PD, progressive disease; SD, stable disease; CI, confidence interval.
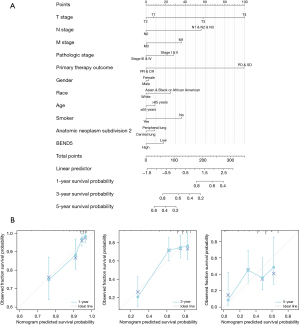
Nomogram plot and calibration plot
To assess the survival probability of patients with LUAD at 1, 3, and 5 years, a nomogram plot (Figure 4A) was constructed incorporating BEND5 expression levels along with independent clinical characteristics. The points for genetic score, age, and TNM stage were added to obtain the total points, with a higher number of total points on the nomogram suggesting a worse prognosis. A calibration curve was established to evaluate the nomogram model’s prediction accuracy. The nomogram-predicted 1-, 3- and 5-year OS was found to match the actual survival results, as indicated by the calibration curves (Figure 4B).
PPI network plotting and heatmap
PPI network analysis, performed using the online STRING database, was used to evaluate the possible relationships of BEND5 with other genes. The PPI network graph includes BEND5 and 10 proteins to which it was strongly related (Figure 5A). The proteins KCNH4, PRSS12, and COL24A1 had the highest combined scores, with corresponding values of 0.647, 0645, and 0.637, respectively.

The heat map expression patterns of genes related to BEND5 in LUAD samples are depicted in Figure 5B. The top 10 genes that were positively associated and the top 10 genes that were negatively associated with BEND5 were identified. The top 10 genes that showed a positive correlation with BEND5 were GPC3, APBB1, AGBL4, FLJ37453, USP51, TMCC2, SCD5, FAM229B, GALNT16, and NDRG2. The top 10 genes that showed a negative correlation with BEND5 were NATD1, TMEM231, PRRT3, C1QTNF2, RAI2, TTC25, GASK1A, TESK2, COLQ, and AL691432.2.
The functional terms enriched for BEND5-correlated genes
Mountain plots depicting the five functional terms that met the criteria of P.adjust <0.05, q value <0.25, and |NES| >1 are shown in Figure 6A. Through GSEA analysis, BEND5-associated genes were revealed to be primarily enriched in two functions: M_PHASE (Figure 6B) and SIGNALING_BY_NUCLEAR_RECEPTORS (Figure 6C).
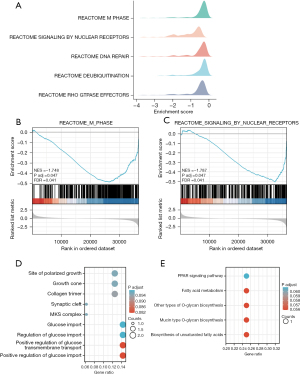
The top 10 genes that were positively and negatively related to BEND5 were found to be significantly enriched in several BP including the site of polarized growth, growth cone, collagen trimer, synaptic cleft, MKS complex, glucose import, regulation of glucose import, positive regulation of glucose transmembrane transport, and positive regulation of glucose import (Figure 6D). Enriched KEGG pathways for the positively and negatively associated genes included the PPAR signaling pathway, fatty acid metabolism, other types of O-glycan biosynthesis, mucin type O-glycan biosynthesis, and biosynthesis of unsaturated fatty acids (Figure 6E).
Increased BEND5 expression inhibits LUAD cell proliferation
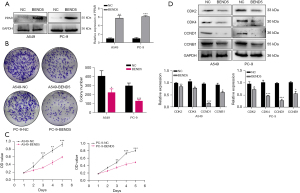
Western blot was applied for the assessment of BEND5 expression levels in five LUAD cell lines and a human bronchial epithelial cell line. As shown in Figure S1A, results revealed that BEND5 had low levels of protein expression in the five different LUAD cell lines, with the A549 and PC-9 cell lines exhibiting the lowest expression levels. This finding prompted us to use these two cell lines for our subsequent research. Western blot analysis revealed that BEND5 plasmids significantly increased BEND5 expression in A549 and PC-9 cells (Figure S1B).
To gain further insight into the intrinsic mechanism of BEND5 in the pathogenesis of LUAD, the PPAR signaling pathway was investigated and was found to be activated by BEND5 overexpression (Figure 7A). Since the PPAR signaling pathway is associated with cell proliferation, in vitro functional assays were performed. Overexpression of BEND5 significantly inhibited A549 and PC-9 cell proliferation, as demonstrated by colonogenic assay and CCK-8 assay results (Figure 7B,7C). Western blot showed that BEND5 overexpression inhibited the cyclin-dependent kinase CDK2/4 and the cyclin proteins CCND1 and CCNB1 (Figure 7D).
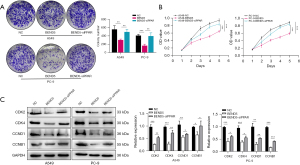
PPAR inhibition restores cell proliferation in BEND5-overexpressing LUAD cells
Previous research shows that overexpression of BEND5 activates the PPAR signaling pathway. However, the contribution of PPAR in the anti-cell proliferation function of BEND5 in LUAD has not yet been clarified. To address this, we conducted colonogenic assay and CCK-8 assays on A549 and PC-9 cells, and the results showed that BEND5 overexpression with PPAR inhibition increased the proliferation ability of LUAD cells (Figure 8A,8B). Results of Western blot further showed that CDK2/4, CCND1, and CCNB1 were restored in BEND5-overexpressing cells with PPAR inhibition (Figure 8C).
Analysis of the relationship between BEND5 expression and immune infiltration in LUAD
Because the functions of BEND5 in the regulation of the immune and tumor microenvironments of LUAD are unknown, the next part of our study investigated the relationship between BEND5 expression and immune scores. The expression of BEND5 was correlated significantly with ESTIMATE and stromal scores in patients with LUAD; however, the immune score did not show a significant correlation (Figure S1C).
Investigating the relationship between BEND5 expression and immune cells in LUAD
BEND5 expression was found to be significantly positively associated with several TIICs, including B cells, DCs, eosinophils, immature DCs, macrophages, mast cells, NK CD56bright cells, NK cells, plasmacytoid DCs, T cells, T follicular helper cells, and T regulatory cells. BEND5 expression was significantly negatively correlated with several others cell types, including NK CD56dim, gamma delta T cells, and Th2 cells (Figure S2A, S2B).
Investigating the relationships between BEND5 expression and 59 ICGs in LUAD
The correlations of BEND5 expression with 23 checkpoint inhibitor genes are shown in Figure S3A. Only 14 of the 23 ICGs showed a statistically significant correlation with BEND5 expression, and the other ICGs had no significant correlation. BEND5 expression was positively correlated with BTLA, CTLA4, SLAMF7, IDO1, ARG1, IL13, IL4, ADORA2A, VEGFB, VTCN1, EDNRB, and IL12A, and negatively correlated with CD274 and VEGFA (Figure S3B).
The correlations of BEND5 expression with 36 checkpoint stimulator genes are shown in Figure S4A. Only 16 of the 36 ICGs showed a statistically significant correlation with BEND5 expression, and the other ICGs had no significant correlation. BEND5 expression was positively correlated with CX3CL1, TNFRSF14, BTN3A1, CD40LG, IL2, CD27, CD28, TNFRSF4, CD40, ENTPD1, SELP, ITGB2, TNF, and CD70, and negatively correlated with IFNG and TNFSF9 (Figure S4B).
Discussion
LUAD is a common respiratory cancer whose morbidity and mortality rates are constantly increasing. As LUAD can develop resistance to radiation and chemotherapy, there is an unmet need for new treatment approaches. To improve the early diagnosis and survival rates of patients with LUAD, specific major molecular pathways and multiple sensitive and reliable biomarkers need to be identified. The relationship between BEND5 and tumor development has been shown in earlier studies (14,15). However, an in-depth description of the role of BEND5 in LUAD is lacking. In the current study, we developed a prognostic model based on BEND5 gene expression in LUAD. Enrichment analysis showed that BEND5 was involved in processes such as cell proliferation and immune infiltration of tumor cells. This is the first investigation to study the effects of BEND5 in LUAD in vitro, and our findings may contribute to the development of future investigations.
BEND5 belongs to the BEN structural domain family, whose members are present in a variety of animal proteins. Despite the fact that the mammalian genome encodes many BEN domain proteins, little research has been done on the BEN domains (10). The first member of this family to be identified was BANP (also known as BEND1 or SMAR1) (28). The tumor suppressor SMAR1 has been reported to act as a transcriptional regulator of the cyclin D1 protein and inhibits its expression (29). In addition, SMAR1 is a protein that interacts with p53, which inhibits tumor progression in vivo as well as delays the cell cycle (30). Another BEN family member, BEND3, is associated with several human malignancies, such as prostate cancer (31), breast cancer (32), and ovarian cancer (33). Also, BEND3 acts as a transcriptional repressor and is involved in transcriptional repression processes through interaction with histone deacetylases and Sall4 (34). The first study of BEND5 was reported in colorectal cancer and showed the contribution of BEND5 hypermethylation to the proliferation of cells, marking its possibility as a prognostic marker (14). BEND5 has also been identified as a tumor suppressor gene associated with growth and metastasis in breast cancer (15). Our bioinformatics analysis showed that BEND5 is expressed at low levels in many cancer types, including colorectal and breast cancer, which is consistent with the findings reported in previous studies. The results from our study based on TCGA data showed that BEND5 was associated with OS in 14 out of 33 cancers, and with relapse-free survival in 10 out of 33 cancers, indicating that BEND5 could be used as a molecular marker for cancer prognosis. The KEGG enrichment analysis demonstrated the involvement of BEND5 in the PPAR signaling pathway, fatty acid metabolism, mucin-type O-glycan biosynthesis, and other processes.
PPARs belong to a class of ligand-dependent transcriptional regulators that are involved in developmental and metabolic processes (35-37). Three isoforms of PPARs have been identified in different species: α, β/δ, and γ. The most common isoform is PPARβ/δ, which is expressed in all tissues and is involved in functions such as cell proliferation and differentiation (38), lipid and glucose metabolism (39), and inhibition of inflammatory processes (40-42). Several studies have suggested the significance of PPAR in tumorigenesis and progression (43,44). For example, studies have indicated a correlation between PPAR proliferation and the rapid recurrence of non-small cell lung cancer (35,36,44), while others have suggested that PPARβ/δ has a protective effect against lung cancer (40). Therefore, we considered that BEND5 may be involved in the proliferation of LUAD cells through the PPAR signaling pathway. In vitro functional assays validated this conjecture, showing that overexpression of BEND5 inhibited the proliferative capacity of LUAD cells, whereas knockdown of PPAR reversed these effects.
The tumor immune microenvironment is crucial to tumor development. Immunotherapy has become an extremely popular treatment for cancers and is also helpful in LUAD. Although a handful of studies have reported the role of BEND5 in some cancers (14,15), the immunomodulatory function of BEND5 in LUAD is not well understood at present. Therefore, our main focus was to evaluate the relationship between the level of BEND5 expression, immune score, and immune cell profile of LUAD. We found that in LUAD, BEND5 expression was positively correlated with several immune infiltrating cells, including B cells, NK cells, and T cells. In the tumor microenvironment, TILs are heterogeneous immune cell populations that produce different immune responses under the influence of different cellular activation mechanisms and cytokines. The synergistic role of immune infiltrating cells is crucial in the anti-tumor response. Moreover, BEND5 expression was found to be associated with 15 non-cancerous cell types as well as 23 checkpoint inhibitor genes and 36 checkpoint stimulator genes, suggesting that it plays a key role in the regulation of the tumor immune microenvironment. Overall, these findings add to our knowledge of the role of BEND5 in tumor immunology and its putative application as a LUAD biomarker.
To evaluate the present study thoroughly, it is important to recognize both its strengths and limitations. The main strength of this study is that it explored the significance of the BEND5 gene in LUAD through a series of bioinformatic analyses. The study explored the correlation of BEND5 expression patterns with clinical variables as well as the prognostic value of BEND5 and its significantly associated genes, KEGG and signaling pathways, and involvement in tumor immunity in LUAD. Furthermore, the role of BEND5 in LUAD proliferation was also validated by in vitro experiments. Our study’s main limitation is the absence of in vivo tests to confirm the involvement of BEND5 in LUAD proliferation. Also, it remains to be seen whether BEND5 regulates LUAD proliferation through multiple signaling pathways.
To the best of our knowledge, our study has revealed, for the first time, the role of BEND5 in the progression and pathogenesis of LUAD, and has identified its potential clinical value. The potential clinical utility of the current study’s primary findings should be underlined. The survival analysis results indicate the potential utility of molecular characterization of patients who are candidates for treatment with BNED5 medicines as a precision medicine methodology. BEND5 agonists have the potential to be a novel therapy strategy for LUAD. Considering the significance of BEND5 to LUAD prognosis, BEND5 may be a meaningful prognostic biomarker. Therefore, it is necessary to explore the role of BEND5 in LUAD and the molecular mechanisms involved more extensively in future.
Conclusions
Using integrated bioinformatics analyses, our study demonstrated that BEND5 was downregulated in LUAD and could potentially serve as a prognostic biomarker for the disease. BEND5 inhibited the proliferation of LUAD cell lines by activating the PPAR signaling pathway. Our results provide evidence for prospective research of the associations of LUAD with the BEND5 gene; however, more investigations are warranted to comprehensively illuminate the role of BEND5 in LUAD.
Acknowledgments
Funding: None.
Footnote
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-314/dss
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-314/prf
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-314/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Bade BC, Dela Cruz CS. Lung Cancer 2020: Epidemiology, Etiology, and Prevention. Clin Chest Med 2020;41:1-24. [Crossref] [PubMed]
- Ruiz-Cordero R, Devine WP. Targeted Therapy and Checkpoint Immunotherapy in Lung Cancer. Surg Pathol Clin 2020;13:17-33. [Crossref] [PubMed]
- Travis WD, Brambilla E, Noguchi M, et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society: international multidisciplinary classification of lung adenocarcinoma: executive summary. Proc Am Thorac Soc 2011;8:381-5. [Crossref] [PubMed]
- Koo HK, Jin SM, Lee CH, et al. Factors associated with recurrence in patients with curatively resected stage I-II lung cancer. Lung Cancer 2011;73:222-9. [Crossref] [PubMed]
- Ganesh K, Massagué J. Targeting metastatic cancer. Nat Med 2021;27:34-44. [Crossref] [PubMed]
- Cheng WC, Chang CY, Lo CC, et al. Identification of theranostic factors for patients developing metastasis after surgery for early-stage lung adenocarcinoma. Theranostics 2021;11:3661-75. [Crossref] [PubMed]
- Zeng H, Chen W, Zheng R, et al. Changing cancer survival in China during 2003-15: a pooled analysis of 17 population-based cancer registries. Lancet Glob Health 2018;6:e555-67. [Crossref] [PubMed]
- Spella M, Stathopoulos GT. Immune Resistance in Lung Adenocarcinoma. Cancers (Basel) 2021;13:384. [Crossref] [PubMed]
- Abhiman S, Iyer LM, Aravind L. BEN: a novel domain in chromatin factors and DNA viral proteins. Bioinformatics 2008;24:458-61. [Crossref] [PubMed]
- Dai Q, Ren A, Westholm JO, et al. The BEN domain is a novel sequence-specific DNA-binding domain conserved in neural transcriptional repressors. Genes Dev 2013;27:602-14. [Crossref] [PubMed]
-
USNLoM NCfBI 2015 . Available online: https://www.ncbi.nlm.nih.gov/gene/79656 - Shi G, Bai Y, Zhang X, et al. Bend family proteins mark chromatin boundaries and synergistically promote early germ cell differentiation. Protein Cell 2022;13:721-41. [Crossref] [PubMed]
- Lin RK, Hung WY, Huang YF, et al. Hypermethylation of BEND5 contributes to cell proliferation and is a prognostic marker of colorectal cancer. Oncotarget 2017;8:113431-43. [Crossref] [PubMed]
- Shi Y, Zhang D, Chen J, et al. Interaction between BEND5 and RBPJ suppresses breast cancer growth and metastasis via inhibiting Notch signaling. Int J Biol Sci 2022;18:4233-44. [Crossref] [PubMed]
- Liu J, Lichtenberg T, Hoadley KA, et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018;173:400-416.e11. [Crossref] [PubMed]
- Pan-cancer analysis of whole genomes. Nature 2020;578:82-93. [Crossref] [PubMed]
- Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519-25. [Crossref] [PubMed]
- Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Yan S, Wong KC. GESgnExt: Gene Expression Signature Extraction and Meta-Analysis on Gene Expression Omnibus. IEEE J Biomed Health Inform 2020;24:311-8. [Crossref] [PubMed]
- Liu Z, Wang J, Li S, et al. Prognostic prediction and immune infiltration analysis based on ferroptosis and EMT state in hepatocellular carcinoma. Front Immunol 2022;13:1076045. [Crossref] [PubMed]
- Yang B, Fan Y, Liang R, et al. Identification of a prognostic six-immune-gene signature and a nomogram model for uveal melanoma. BMC Ophthalmol 2023;23:2. [Crossref] [PubMed]
- Li D, Shi Z, Liu X, et al. Identification and development of a novel risk model based on cuproptosis-associated RNA methylation regulators for predicting prognosis and characterizing immune status in hepatocellular carcinoma. Hepatol Int 2023;17:112-30. [Crossref] [PubMed]
- The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013;45:580-5. [Crossref] [PubMed]
- Keen JC, Moore HM. The Genotype-Tissue Expression (GTEx) Project: Linking Clinical Data with Molecular Analysis to Advance Personalized Medicine. J Pers Med 2015;5:22-9. [Crossref] [PubMed]
- Thorsson V, Gibbs DL, Brown SD, et al. The Immune Landscape of Cancer. Immunity 2019;51:411-2. [Crossref] [PubMed]
- Wu L, Jie B. Protumor Effects of Histone H3-H4 Chaperone Antisilencing Feature 1B Gene on Lung Adenocarcinoma: In Silico and In Vitro Analyses. Comput Math Methods Med 2021;2021:5005459. [Crossref] [PubMed]
- Birot A, Duret L, Bartholin L, et al. Identification and molecular analysis of BANP. Gene 2000;253:189-96. [Crossref] [PubMed]
- Rampalli S, Pavithra L, Bhatt A, et al. Tumor suppressor SMAR1 mediates cyclin D1 repression by recruitment of the SIN3/histone deacetylase 1 complex. Mol Cell Biol 2005;25:8415-29. [Crossref] [PubMed]
- Kaul R, Mukherjee S, Ahmed F, et al. Direct interaction with and activation of p53 by SMAR1 retards cell-cycle progression at G2/M phase and delays tumor growth in mice. Int J Cancer 2003;103:606-15. [Crossref] [PubMed]
- Hyytinen ER, Saadut R, Chen C, et al. Defining the region(s) of deletion at 6q16-q22 in human prostate cancer. Genes Chromosomes Cancer 2002;34:306-12. [Crossref] [PubMed]
- Orphanos V, McGown G, Hey Y, et al. Proximal 6q, a region showing allele loss in primary breast cancer. Br J Cancer 1995;71:290-3. [Crossref] [PubMed]
- Orphanos V, McGown G, Hey Y, et al. Allelic imbalance of chromosome 6q in ovarian tumours. Br J Cancer 1995;71:666-9. [Crossref] [PubMed]
- Sathyan KM, Shen Z, Tripathi V, et al. A BEN-domain-containing protein associates with heterochromatin and represses transcription. J Cell Sci 2011;124:3149-63. [Crossref] [PubMed]
- Sun X, Ritzenthaler JD, Zhong X, et al. Nicotine stimulates PPARbeta/delta expression in human lung carcinoma cells through activation of PI3K/mTOR and suppression of AP-2alpha. Cancer Res 2009;69:6445-53. [Crossref] [PubMed]
- Genini D, Garcia-Escudero R, Carbone GM, et al. Transcriptional and Non-Transcriptional Functions of PPARβ/δ in Non-Small Cell Lung Cancer. PLoS One 2012;7:e46009. [Crossref] [PubMed]
- Gamdzyk M, Doycheva DM, Malaguit J, et al. Role of PPAR-β/δ/miR-17/TXNIP pathway in neuronal apoptosis after neonatal hypoxic-ischemic injury in rats. Neuropharmacology 2018;140:150-61. [Crossref] [PubMed]
- Hojka A, Rapak A. Peroxisome proliferator-activated receptors (PPAR). Antiproliferative properties. Postepy Hig Med Dosw (Online) 2011;65:404-13. [Crossref] [PubMed]
- Chen J, Montagner A, Tan NS, et al. Insights into the Role of PPARβ/δ in NAFLD. Int J Mol Sci 2018;19:1893. [Crossref] [PubMed]
- Lakshmi SP, Reddy AT, Banno A, et al. PPAR Agonists for the Prevention and Treatment of Lung Cancer. PPAR Res 2017;2017:8252796. [Crossref] [PubMed]
- Sng MK, Chan JSK, Teo Z, et al. Selective deletion of PPARβ/δ in fibroblasts causes dermal fibrosis by attenuated LRG1 expression. Cell Discov 2018;4:15. [Crossref] [PubMed]
- Banno A, Reddy AT, Lakshmi SP, et al. PPARs: Key Regulators of Airway Inflammation and Potential Therapeutic Targets in Asthma. Nucl Receptor Res 2018;5:101306. [Crossref] [PubMed]
- Wang X, Wang G, Shi Y, et al. PPAR-delta promotes survival of breast cancer cells in harsh metabolic conditions. Oncogenesis 2016;5:e232. [Crossref] [PubMed]
- Müller R. PPARβ/δ in human cancer. Biochimie 2017;136:90-9. [Crossref] [PubMed]
(English Language Editor: J. Reylonds)


