
Integrative learning in developing an immunologic lncRNA signature as a consensus risk-stratification tool for lung adenocarcinoma
Highlight box
Key findings
• ILLS demonstrated superior and stable prognostic prediction ability and thus has potential as a tool for assisting in risk classification and clinical decision-making in patients with LUAD.
What is known and what is new?
• Multidisciplinary treatments like targeted therapy and immunotherapy appear to be effective in only a few patients with LUAD. resulting in insubstantial development of an overall prognosis.
• To guarantee the robust prognostic prediction and generalizability of ILLS, 101 compositions of algorithms were adopted to fit the best model which was cross-domain–validated in 4 GEO data sets and then transversely compared to 49 published signatures.
What is the implication, and what should change now?
• This ILLS model can help to predict prognosis and benefit immunotherapy by personalizing treatment for patients with LUAD. Further high-quality preclinical studies should be conducted to clarify the mechanism and collect more evidence to confirm these findings.
Introduction
It is estimated that over 350 people die from lung cancer every day, which is more than from prostate, pancreatic, and breast cancers combined, and two-and-a-half times more than from colorectal cancer, the second largest cause of cancer-related death. Lung adenocarcinoma (LUAD) has become the most prevalent lung cancer subtype and has increased in frequency (1,2). Although remarkable advances have been made in the multidisciplinary treatment of LUAD, the unprecedented effectiveness has been observed in only a small portion of patients. Targeted therapy is limited to histological subtypes containing operable driver mutations, and durable responses to immunotherapy are not common, resulting in the insubstantial development of an overall prognosis for patients with LUAD (3,4). Therefore, the exploration of new prognostic biomarkers should be emphasized to stratify risk and provide optimal treatment for patients with LUAD. Unlike proteins, the stability of cancer-related Long noncoding RNA (lncRNA) is more suitable for cancer screening and detection. Although the research of circulating lncRNA is still in its early stage, the interest in lncRNA is growing worldwide and new technologies are emerging to develop potential (5). In LUAD, a significant correlation has been observed between tumor genomic features and the immune microenvironment. The immune microenvironment is highly heterogeneous between and within patients, and even in distinct regions of the same tumor, nearly a third of the samples show varying degrees of immune infiltration (6). LncRNAs have attracted increasing attention as biomarkers for cancer diagnosis and therapy as well as therapeutic targets due to their abnormal regulation in the process of tumorigenesis. Several experiments have demonstrated that lncRNA plays crucial roles in both cancer and the immune response through multiple regulatory mechanisms (7,8). Adjacent immune cells and various types of cells in the tumor immune microenvironment (TIME) continuously interact and evolve, affecting intrinsic and extrinsic processes such as proliferation, invasion, epithelial-mesenchymal transition, angiogenesis, and drug resistance (9). For example, He et al. focused on identifying immune-associated lncRNA in LUAD and found its prediction value in immune response between tumor and normal tissue (10). Gong et al. revealed the role of immune-related competing endogenous RNA (ceRNA) network in LUAD, helping to explore novel target of anti-tumor immunotherapy (11).
In this study, we mined public data from The Cancer Genome Atlas (TCGA) and the Gene Expression Omnibus (GEO) databases, identified immune-related lncRNAs, and developed an immunologic lncRNA signature (ILLS) with reliable prognostic value with the aim of stratifying risk and predicting prognosis. In addition, drug sensitivity analysis was conducted to provide novel insights into personalized treatment decisions with the aim of standardizing the prediction model reporting process and reporting quality evaluation. We present the following article in accordance with the TRIPOD reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-372/rc).
Methods
Obtaining and processing the public data
Raw data of patients with LUAD were obtained from TCGA (https://cancergenome.nih.gov/) and GEO databases (https://www.ncbi.nlm.nih.gov/geo/) and included RNA-sequencing (RNA-seq) profiling and clinical information. In this study, TCGA-LUAD data sets were used in supervised learning to construct our required signature. In addition, GSE50081, GSE31210, GSE37745, and GSE30219, 4 independent data sets, were adopted as validation sets to evaluate the generalizability of the signature. Only patients with complete expression profiles, overall survival (OS) information, and necessary available clinical data were included for analysis. A total of 1,140 patients with LUAD, RNA expression matrix, and complete OS information were included across all the data sets. The Imvigor210 data set with response data was retrieved to assess the ability of ILLS to predict immunotherapy efficacy. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
The raw read count of RNA-seq from TCGA-LUAD data set was acquired in a unit of transcripts per kilobase million. In the GEO database, all files were received from the platform GPL570, and the “Affy” package running in R software (version 4.2.1) facilitated log-2 transformation through the robust multiarray averaging method for the above original data, in a process known as normalization. Via gene annotations from the Homo sapiens GRCh38, we found 16,773 lncRNAs in the TCGA-LUAD data sets. We reannotated the probe set of the gene GPL570 array and converted it into 1,134 lncRNAs.
Consensus clustering
Single-sample gene set enrichment analysis was employed to quantify the infiltration degree of 28 immune cells through use of the “GSVA” package in R software on the TCGA-LUAD data set. Subsequently, based on the infiltration score of diverse immune cells, consensus clustering was used to discover an appropriate cluster. The procedure was implemented via the “ConsensusClusterPlus” package in R. Next, the proportion from the ambiguous clustering (PAC) score and k-means algorithm was consistently applied to determine the best number of clusters. The micro- and the macrolevels of immune infiltration difference were visualized in a boxplot and heat map.
Weighted correlation network analysis (WGCNA)
By means of R software’s “WGCNA” package, lncRNA coexpression networks were generated in TCGA-LUAD data set. In addition to the consequence of clustering, other clinical traits influencing clinical decision-making including age, sex, stage defined by American Joint Committee on Cancer (AJCC), pathological TNM stage, tumor mutation burden (TMB), microsatellite instability (MSI), and neoantigen load (NAL) state were also factored into the calculation. To satisfy the criterion of approximate scale-free topology, a suitable β was calculated. Moreover, a topological overlap matrix (TOM) was transformed from the weighted adjacency matrix, and then corresponding dissimilarity was generated, termed “1-TOM”. To recognize those module eigengenes significantly correlated with cluster traits, the eigengenes with key modules that had the highest correlations were extracted. Only hub lncRNAs that met the criteria of gene significance >0.5 and module membership >0.6 were defined as immune-related lncRNAs.
ImmLnc enrichment analysis
ImmLnc is a web-based resource for investigating the immune-related pathways of lncRNAs, and the process is achieved with R package “ImmLncRNA” containing 3 functions. According to all messenger RNA (mRNA) expression profiles, the tumor purity of every sample was first estimated with the ESTIMATE algorithm. The partial correlation coefficient and P value between genes and lncRNAs were then calculated. Based on the gene set enrichment analysis, the enrichment score, P value, and the combined lncRES, statistics were obtained to identify the potential lncRNA modulators of immune-related pathways. Among them, only those with lncRES >0.995 and false-discovery rate (FDR) <0.05 were considered statistically significant (12). Previous literature suggests that if a particular lncRNA acts as a crucial immunomodulator, its relevant genes ought to enrich in the high or low placing of immune-related pathways (13). The lncRNAs corresponding to the top 5 pathways were selected for further research.
Integrative approach-based ILLS development
Based on the intersection of WGCNA and ImmLnc results, univariate Cox analysis was used to determine if the prognostic lncRNAs correlated with OS in TCGA-LUAD data set. In order to develop signatures with superior and stable predictive ability, by means of integrative processing, we applied 10 algorithms and 101 combinations for developing our desired signature. We included popular algorithms, such as least absolute shrinkage and selection operator (LASSO), ridge, elastic network, supervised principal components, and random survival forest, as well as relatively uncommon but equally powerful ones such as generalized boosted regression modelling, survival support vector machine, CoxBoost, and partial least squares regression for Cox. Cross-validation in the leave-one-out principle was used to calculate the concordance index (C-index) for the 4 validation data sets (GSE31210, GSE37745, GSE30219, and GSE50081). The constructed model with the highest mean C-index was considered optimal.
Statistical analysis
All survival statistics computing and results illustration were performed using R software (version 4.2.1). The relevant coefficient and corresponding risk score of each LUAD sample were calculated. We distinguished the high-risk group from the low-risk group according to the median risk score (i.e., the optimal cutoff value). Between the high- and low-risk groups, Kaplan-Meier curve analysis was performed to show the OS difference of patients with LUAD. Within the range of possible values of a predictive variable, we compared the area under the receiver operating characteristic (ROC) curve (AUC) to assess the diagnostic accuracy of ILLS, and the time-dependent AUC for survival variables was conducted in 1-, 3-, and 5-year survival. We also calculated the Akaike information criterion value to locate the cutoff point for the AUC of 1 year. Through univariate and multivariate Cox regression, ILLS was tested to determine whether it was an independent prognostic factor. The same approaches were applied to the 4 validation data sets (GSE31210, GSE37745, GSE30219, and GSE50081) for cross-domain validations, which helped to prove the reliability of ILLS. The common R packages used in the above validation included “survival”, “survminer,” “timeROC”, “ggplot2”, “limma”, “scatterplot3d”, “ggpubr”, and “Rtsne”.
Transverse comparison of the published lncRNA signatures for LUAD
With advances in high-throughput sequencing and big-data analysis techniques, there has been explosive increase in studies exploring effective predictors and therapeutic targets. In this paper, we reviewed the lncRNA signatures published in the MEDLINE database. The retrieved articles related to the prognosis analysis for patients with LUAD were included, and then we conducted a transverse comparison to rank their performance against ILLS.
Usability research
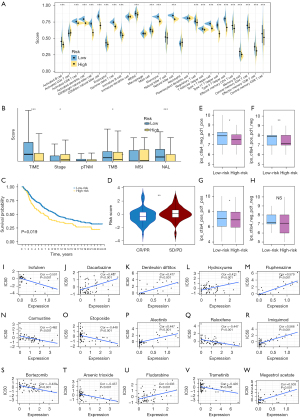
We classified 28 immune cell infiltrations into high- and low-risk groups and determined which subset increased most significantly. AJCC stage, pathological TNM stage, and several emerging immunotherapy markers (i.e., TMB, NAL, and MSI) were also used to determine whether their status was consistent with the desired risk and further ensured their potential as candidate Additionally, 2 data sets were used to help verify the ability of ILLS to predict the immunotherapeutic benefits. Moreover, the gene expression and immunotherapy response files were also retrieved from the IMvigor210 data set, and the differences in prognosis and response of the model constructed by the ILLS was determined. Subsequently, using The Cancer Immunome Atlas (https://www.tcia.at/home), we downloaded the immunophenoscore (IPS) of TCGA-LUAD patients, which mainly refers to the major histocompatibility complex (MHC) molecules, effector cells, immune checkpoints, and immunosuppressive cells. The IPS was confirmed as having a positive correlation with tumor immunogenicity. The immunogenicity was quantified on a scale from 0 to10 based on the gene expression in the representative cell types, thereby reflecting the immunotherapy response. For suiting chemotherapy to the needs of the case, we obtained the top 15 drugs with the most significant correlations by performing a correlation analysis between half-maximal inhibitory concentration (IC50) values and ILLS expression, which is the widely used method for cancer drug-testing databases. CellMiner (NCI-60; https://discover.nci.nih.gov/cellminer) contributed significantly to this exploration by providing compound activity information and RNA-seq expression.
Results
Sample clustering and key immune differentiator findings
Based on the infiltration score of 28 separate immune cells deduced by single-sample gene set enrichment analysis in TCGA-LUAD samples, cluster analysis indicated typing into 2 distinct clusters (Figure 1A). The proportion statistic of ambiguous clustering displayed the fittest cluster number = 2, k-means clustering also achieved the same result (Figure 1B,1C). We then identified a clustering trait which classified C1/C2 into immune-cold or immune-hot subtypes, and a difference was indeed found (Figure 1D,1E) which was then incorporated into the construction of a coexpression network. Seven modules with individual colors were identified. The highest correlation and significance were observed between the yellow module and the clustering trait (Figure 1F). Among the eigengenes of the yellow module, 589 hub lncRNAs with satisfactory gene significance and module membership were taken as immune-related lncRNAs, presenting the strongest correlation in the classification of immune subtypes.
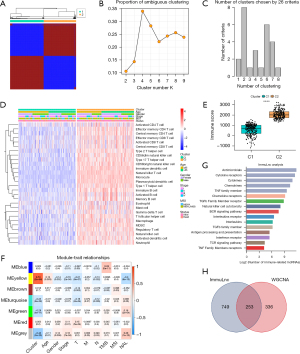
Immune pathway regulator generation
In all, 1,002 lncRNAs met the significance threshold and were selected and determined to enrich the top 5pathways of antimicrobials, cytokine receptors, cytokines, chemokines, and tumor necrosis family (TNF) family members (Figure 1G). Finally, we extracted 253 immune-related lncRNAs that overlapped in the WGCNA and ImmLnc results for further development (Figure 1H).
Consensus signature development
Among the above immune-related lncRNAs, 52 prognostic lncRNAs were identified in the univariate Cox analysis. Comprehensive approaches were then employed to develop a consensus signature. According to the model performance in the GSE31210, GSE37745, GSE30219, and GSE50081 data sets, among the 101 algorithm compositions, the model constructed by LASSO and stepwise Cox in both directions demonstrated a superior accuracy and generalizability. The average C-index reached a high of 0.69 (Figure 2A,2B). The ILLS was as follows: LINC01138, LPP-AS2, LINC00857, UCA1, and LINC01116, with positive coefficients regarded as risk factors. Conversely, GAS6-AS1, AF186192.1, NNT-AS1, and LINC00996 were negatively coefficient and thus considered to exert a protective role (Figure 2C). From the division into high- and low-risk groups, survival analysis showed that patients from the former experienced lower OS than the latter in TCGA-LUAD (P<0.0001), GSE31210 (P<0.0001), GSE30219 (P<0.0001), GSE50081 (P=0.009), GSE37745 (P=0.023) and the Meta-Datasets (P<0.0001) (Figure 2D-2I).
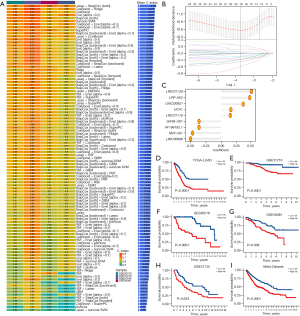
ILLS performance testing
To prioritize sensitivity and specificity, we tested the distinguishing capacity of ILLS, and revealed that the 1-, 3-, and 5-year AUCs were 0.712, 0.711, and 0.653 in TCGA-LUAD data set; 0.886, 0.644, and 0.683 in the GSE31210 data set; 0.785, 0.596, and 0.634 in the GSE30219 data set; 0.689, 0.547, and 0.563 in the GSE50081 data set; and 0.751, 0.705, and 0.617 in the GSE37745 data set, respectively (Figure 3A-3E). The best cutoff points were assessed to divide patients into high- and low-risk groups (Figure 3F-3J). Moreover, ILLS was found to be an independent prognostic factor in TCGA-LUAD, GSE31210, GSE30210, and GSE50081 data sets. Although ILLS had a certain degree of predictive value for OS in the GSE37745 data set, it was not found to be an independent prognostic factor (Figure 3K-3O), possibly due to the small sample size. We found 49 recently published articles focused on developing a lncRNA-based signature for risk stratification and therapeutic assessment in patients with LUAD. The signatures examined were associated with several key bioprocesses, including immune microenvironments, N6-methyladenosine, pyroptosis, ferroptosis, hypoxia, autophagy, and redox genome instability, among others. Their wide-range predictive ability was compared using C-index statistics. The results demonstrated that ILLS possessed desirable discriminative ability and generalizability among these signatures. It is worth noting that most of signatures, worked very well in their initial training model and a small portion of validation data sets, but performed poorly in other data sets (Figure 4).
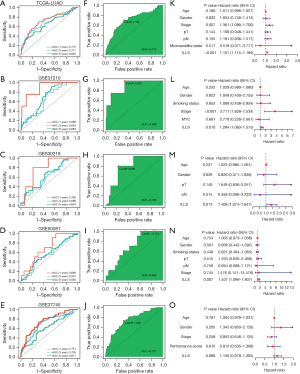
Clinical guidance
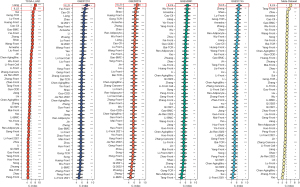
Correlation analysis between the low- and high-risk groups of the 28 immune cell subtypes revealed the expected outcome. Except for activated CD4 T cells, immature dendritic cells, macrophages, natural killer T cells, neutrophils and plasmacytoid dendritic cells, all the other subtypes (22 subtypes) of immune cells displayed significantly higher infiltration levels in the tumor site of the patients with low risk (P<0.001) (Figure 5A). The TIME was not the only strong correlation deduced from the model, and differences in AJCC stage, TMB, and NAL were also identified. In the patients with high risk, TIME and NAL tended to show a higher degree of correlation, while AJCC stage and TMB showed a lower level of correlation; however, differences in pathological TNM stage and MSI were nonsignificant (Figure 5B). According to the genetic and clinical information of the IMvigor210, the high-risk group tended to possess a worse survival in the model (Figure 5C). As the risk score increased, the proportion of patients presenting with stable disease or progressive disease increased, and patients at low risk were more likely to have a complete or partial response status (P<0.01) (Figure 5D). Overall, these results indicate that ILLS can, to a degree, be an alternative immunotherapy biomarker and assist in selecting patients sensitive to immune checkpoint inhibitors (ICIs). Immunogenicity quantification suggested that for PD-1/PD-L1/PD-L2, CTLA4, and PD1/PD-L1/PD-L2 plus CTLA4 blockers, an ideal immunotherapy benefit could be achieved in the low-risk group, especially for CTLA-4 blockers (P<0.01), while for the total IPS, there was no significant response difference between the 2 groups (Figure 5E-5H). Pharmacological analysis revealed 15 chemotherapeutic medicines that showed a high correlation between ILLS expression and drug sensitivity. The higher the expression of ILLS, the more sensitive tumor cells were to fluphenazine, imiquimod, megestrol acetate, dacarbazine, denileukin diftitox, hydroxyurea, alectinib, and fludarabine as feasible alternative treatments. As ILLS expression increased, irofulven, carmustine, etoposide, raloxifene, bortezomib, arsenic trioxide, and trametinib appeared to be less effective in patients with LUAD (all P values <0.001) (Figure 5I-5W).
Discussion
LUAD is characterized by high interpatient and intratumor heterogeneity, and multiple gene mutations and epigenetic changes drive the tumor development and progression. It has been identified that within the TIME, oncogenic processes and histopathology heterogeneity are main drivers for tumor evolution (14-16). Intratumor lncRNA also leads to diverse TIME (7). As there is a lack of accurate predictors concerning the effect of continuous treatment, we conducted this study in order to derive a consensus signature based on TIME characteristics with the added aim of accurately identifying high-risk groups to facilitate clinical decision-making.
In this study, RNA-seq profiles and clinical information were mined from publicly available data sets. Depending on the immune status of samples from TCGA-LUAD data set, we successively used clustering analysis, WGCNA, and the integrated ImmLnc algorithm, and then combined the extracted immune-related lncRNA with survival data. Via integrative approaches, we ultimately derived 9 lncRNAs for the consensus ILLS. After repeated cross-domain validation and transverse comparison, the stability and superiority of ILLS was confirmed. However, with treatment being the correct solution, in the end, testing the ability for immunotherapy response prediction and discovering potential therapeutic agents can assist in bringing fresh insight into tailoring regiments for patients at high risk.
The lncRNAs can function as ceRNAs by competitively binding to shared sequences of microRNAs (miRNAs) and affecting the expression levels of their downstream mRNA, Reducing/enhancing its expression, which form lncRNA-miRNA-mRNA interactions or ceRNA networks. This dual relationship is important in affecting proliferation and metastasis of cancer cells (17,18).
LINC01138 has proven to be a promising prognostic indicator via the LINC01138-PRMT5 axis in hepatocellular carcinoma and was concluded to be a risk factor driving malignancies in patients with LUAD (19,20). Through in vivo and in vitro assays, LPP-AS2 was assessed to be an oncogene to modulate the downstream protein of EGFR, promoting glioma tumorigenesis via a miR-7-5p/EGFR/PI3K/AKT/c-MYC feedback loop; likewise, a higher expression of LPP-AS2 was found to be associated with a poorer prognosis in LUAD (21,22). In addition, lncRNA LINC00857 was shown to have a prominent role in lung cancer progression. For example, silencing of LINC00857 can mediate the proliferation of lung cancer cells through inducing cell apoptosis and autophagy (23). In recent years, accumulating research has demonstrated the oncogenic role of lncRNA UCA1 in a wide array of human cancer cell lines and patient samples, including those of LUAD (24). A high index of LINC01116 expression was associated with gefitinib resistance in non-small cell lung cancer (NSCLC) and unfavorable outcomes (25). GAS6-AS1 was also shown to serve as a tumor suppressor in LUAD, mainly regulating the reprogramming of glucose metabolism (26). AF186192.1-associated cancer research has only been conducted in LUAD and has been identified as a methylation driving mechanism (27). An in-depth study showed that the regulatory mechanism of NNT-AS1 is as a miRNA sponge, critically contributing to the proliferation, invasion, metastasis, and apoptosis of lung cancer cells (28). Within the immune-editing theory approach, lncRNA LINC00996 was identified as a potential therapeutic target in LUAD (29). In the present study, the most highly expressed biomarkers in the ILLS—whether acting as oncogenes or antioncogenes in tumor progression—consistently followed the expected prognostic trends in accordance with the general findings of basic experiments. It is concerning that although some researchers have already engaged in the investigation of biomarkers with diverse depth and scope, little is known about their immunity and immunotherapy. It is hoped that more relevant explorations will be carried out in this high-potential area in the future.
Despite frequent clinical application, ICI therapy still produces a limited response rate, and thus achieving the accurate recognition of target patients is highly desirable. In the context of immunotherapy, ILLS can contribute to identifying those patients with satisfactory response, which facilitates clinical guiding in applying anti-PD-1 and anti-CTLA4 drug to a certain degree. Functional genomic experiments have suggested that LUAD and urothelial carcinoma of the bladder have a similar distribution of certain immune cells, such as macrophages, with the same survival trend occurring in patients with high a proportion CD8+ T cells and a low proportion of macrophages (30,31). There also have confirmed common immunomodulatory molecules and similar patterns in overall immune response between them. Therefore, we tried to verify the efficacy prediction of ILLS for immunotherapy through the IMvirgor210 data set with the aim of providing a reference for preclinical studies (32,33). In general, ILLS demonstrated the ability to predict immunotherapy in terms of immunogenicity and response results. Up to now, drug resistance remains a huge challenge in the reason of causing therapeutic failure (34). The effect of some immunosuppressors such as Carmustine, Dacarbazine, Etoposide can also be predicted based on ILLS expression.
The immune infiltration within TME was increasingly recognized to be associated with immunotherapy response and patient prognosis among LUAD and other cancers (35). Within TIME, the crosstalk between tumor and infiltrating immune cells is active and involves the whole process of immunotherapy (36). In NSCLC, a meta-analysis summarized that the CD8+ T cell subtype was the best predictor of survival (37). CD4+ T helper cells promote the priming and effector and memory functions of CD8+ cytotoxic T cells (38). Tumor-associated macrophages display suppressive activity to blunt CD8+ T cells (39). Dendritic cells are powerful antigen-presenting cells that are critical for initiating T-cell responses (40). Natural killer cells are crucial for immunosurveillance, and higher susceptibility to cancer and metastasis arises when they are diminished (41). The desired distribution of infiltrating immune cells between the high- and low-risk groups in this study can explain this consistency of the immunotherapy in previous findings.
Several large prospective clinical trials have found that patients with high TMB levels contributed to improving response to immunotherapy and survival, and this observation of benefit extended to cases with all levels of PD-L1 expression (42-44). Multi-aspect analyses are beneficial in fostering a better understanding of the mechanisms of ILLS that are essential for follow-up studies and clinical guidance. Whole-exome sequencing has clarified the relationship between overall TMB/NAL and ICI response in NSCLC (45). TMB and NAL have been strongly correlated, with high TMB tumors co-occurring with increased levels of immunogenic neoantigens; however, only a handful of lung cancers are MSI-high and have DNA mismatch repair (46-48). Moreover, patients with near-high–MSI have been found to have high TMB, harboring higher NAL originating from an elevated genome-wide mutation rate (49,50). In short, the role of MSI in lung cancer is less certain, but a high TMB and NAL tend to be associated with the desired response to ICIs (51), which is in line with the findings our study.
Some limitations to this study should be noted. First, we employed a retrospective study, and although several measures were taken to guarantee the clinical significance of ILLS, further experimental data are needed to support our findings, which presently may not be enough to attract clinical translation and implementation. Second, the relationship between ILLS and immunotherapy is complex. The available public data sets were insufficient to allow us to clarify the relationship between ILLS and immunotherapy. Third, the mechanism of immune function relating to tumorigenesis and treatment is unclear, and, although our understanding of the immune cellular infiltration within TIME used to derive the ILLS is not comprehensive, the result was satisfactory.
Conclusions
Following a mass of bioinformatics computing and algorithm deduction, we derived a superior and stable tool, the ILLS. This ILLS model can help to predict the prognosis and enhance immunotherapy by personalizing treatment for patients with LUAD.
Acknowledgments
Funding: This work was supported by grants from the Guangdong Medical Science and Technology Research Fund Projects (grant No. A2022177).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-372/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-372/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA Cancer J Clin 2022;72:7-33. [Crossref] [PubMed]
- Nicholson AG, Tsao MS, Beasley MB, et al. The 2021 WHO Classification of Lung Tumors: Impact of Advances Since 2015. J Thorac Oncol 2022;17:362-87. [Crossref] [PubMed]
- Miller KD, Nogueira L, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin 2019;69:363-85. [Crossref] [PubMed]
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553:446-54. [Crossref] [PubMed]
- Badowski C, He B, Garmire LX. Blood-derived lncRNAs as biomarkers for cancer diagnosis: the Good, the Bad and the Beauty. NPJ Precis Oncol 2022;6:40. [Crossref] [PubMed]
- Rosenthal R, Cadieux EL, Salgado R, et al. Neoantigen-directed immune escape in lung cancer evolution. Nature 2019;567:479-85. [Crossref] [PubMed]
- Du Z, Fei T, Verhaak RG, et al. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol 2013;20:908-13. [Crossref] [PubMed]
- Park EG, Pyo SJ, Cui Y, et al. Tumor immune microenvironment lncRNAs. Brief Bioinform 2022;23:bbab504. [Crossref] [PubMed]
- Binnewies M, Roberts EW, Kersten K, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med 2018;24:541-50. [Crossref] [PubMed]
- He C, Yin H, Zheng J, et al. Identification of immune-associated lncRNAs as a prognostic marker for lung adenocarcinoma. Transl Cancer Res 2021;10:998-1012. [Crossref] [PubMed]
- Gong WJ, Zhou T, Wu SL, et al. A novel immune-related ceRNA network that predicts prognosis and immunotherapy response in lung adenocarcinoma. Ann Transl Med 2021;9:1484. [Crossref] [PubMed]
- Li Y, Jiang T, Zhou W, et al. Pan-cancer characterization of immune-related lncRNAs identifies potential oncogenic biomarkers. Nat Commun 2020;11:1000. [Crossref] [PubMed]
- Liu Z, Liu L, Weng S, et al. Machine learning-based integration develops an immune-derived lncRNA signature for improving outcomes in colorectal cancer. Nat Commun 2022;13:816. [Crossref] [PubMed]
- Campbell JD, Alexandrov A, Kim J, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 2016;48:607-16. [Crossref] [PubMed]
- Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Tavernari D, Battistello E, Dheilly E, et al. Nongenetic Evolution Drives Lung Adenocarcinoma Spatial Heterogeneity and Progression. Cancer Discov 2021;11:1490-507. [Crossref] [PubMed]
- Zhao M, Feng J, Tang L. Competing endogenous RNAs in lung cancer. Cancer Biol Med 2021;18:1-20. [Crossref] [PubMed]
- Ashrafizadeh M, Gholami MH, Mirzaei S, et al. Dual relationship between long non-coding RNAs and STAT3 signaling in different cancers: New insight to proliferation and metastasis. Life Sci 2021;270:119006. [Crossref] [PubMed]
- Li Z, Zhang J, Liu X, et al. The LINC01138 drives malignancies via activating arginine methyltransferase 5 in hepatocellular carcinoma. Nat Commun 2018;9:1572. [Crossref] [PubMed]
- Xiao L, Huang Y, Li Q, et al. Identification of a prognostic classifier based on EMT-related lncRNAs and the function of LINC01138 in tumor progression for lung adenocarcinoma. Front Mol Biosci 2022;9:976878. [Crossref] [PubMed]
- Zhang X, Niu W, Mu M, et al. Long non-coding RNA LPP-AS2 promotes glioma tumorigenesis via miR-7-5p/EGFR/PI3K/AKT/c-MYC feedback loop. J Exp Clin Cancer Res 2020;39:196. [Crossref] [PubMed]
- Zhuang J, Chen Z, Chen Z, et al. Construction of an immune-related lncRNA signature pair for predicting oncologic outcomes and the sensitivity of immunosuppressor in treatment of lung adenocarcinoma. Respir Res 2022;23:123. [Crossref] [PubMed]
- Su W, Wang L, Zhao H, et al. LINC00857 Interacting with YBX1 to Regulate Apoptosis and Autophagy via MET and Phosphor-AMPKa Signaling. Mol Ther Nucleic Acids 2020;22:1164-75. [Crossref] [PubMed]
- Ghafouri-Fard S, Taheri M. UCA1 long non-coding RNA: An update on its roles in malignant behavior of cancers. Biomed Pharmacother 2019;120:109459. [Crossref] [PubMed]
- Wang H, Lu B, Ren S, et al. Long Noncoding RNA LINC01116 Contributes to Gefitinib Resistance in Non-small Cell Lung Cancer through Regulating IFI44. Mol Ther Nucleic Acids 2020;19:218-27. [Crossref] [PubMed]
- Luo J, Wang H, Wang L, et al. lncRNA GAS6-AS1 inhibits progression and glucose metabolism reprogramming in LUAD via repressing E2F1-mediated transcription of GLUT1. Mol Ther Nucleic Acids 2021;25:11-24. [Crossref] [PubMed]
- Li R, Yang YE, Yin YH, et al. Methylation and transcriptome analysis reveal lung adenocarcinoma-specific diagnostic biomarkers. J Transl Med 2019;17:324. [Crossref] [PubMed]
- Zhou C, Duan S. The Role of Long Non-Coding RNA NNT-AS1 in Neoplastic Disease. Cancers (Basel) 2020;12:3086. [Crossref] [PubMed]
- Yan T, Ma G, Wang K, et al. The Immune Heterogeneity Between Pulmonary Adenocarcinoma and Squamous Cell Carcinoma: A Comprehensive Analysis Based on lncRNA Model. Front Immunol 2021;12:547333. [Crossref] [PubMed]
- Ju M, Bi J, Wei Q, et al. Pan-cancer analysis of NLRP3 inflammasome with potential implications in prognosis and immunotherapy in human cancer. Brief Bioinform 2021;22:bbaa345. [Crossref] [PubMed]
- Varn FS, Wang Y, Mullins DW, et al. Systematic Pan-Cancer Analysis Reveals Immune Cell Interactions in the Tumor Microenvironment. Cancer Res 2017;77:1271-82. [Crossref] [PubMed]
- Ahmed KM, Veeramachaneni R, Deng D, et al. Glutathione peroxidase 2 is a metabolic driver of the tumor immune microenvironment and immune checkpoint inhibitor response. J Immunother Cancer 2022;10:e004752. [Crossref] [PubMed]
- Lim YW, Chen-Harris H, Mayba O, et al. Germline genetic polymorphisms influence tumor gene expression and immune cell infiltration. Proc Natl Acad Sci U S A 2018;115:E11701-10. [Crossref] [PubMed]
- Alfarouk KO, Stock CM, Taylor S, et al. Resistance to cancer chemotherapy: failure in drug response from ADME to P-gp. Cancer Cell Int 2015;15:71. [Crossref] [PubMed]
- Varn FS, Tafe LJ, Amos CI, et al. Computational immune profiling in lung adenocarcinoma reveals reproducible prognostic associations with implications for immunotherapy. Oncoimmunology 2018;7:e1431084. [Crossref] [PubMed]
- Mao X, Xu J, Wang W, et al. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: new findings and future perspectives. Mol Cancer 2021;20:131. [Crossref] [PubMed]
- Zeng DQ, Yu YF, Ou QY, et al. Prognostic and predictive value of tumor-infiltrating lymphocytes for clinical therapeutic research in patients with non-small cell lung cancer. Oncotarget 2016;7:13765-81. [Crossref] [PubMed]
- Borst J, Ahrends T, Bąbała N, et al. CD4(+) T cell help in cancer immunology and immunotherapy. Nat Rev Immunol 2018;18:635-47. [Crossref] [PubMed]
- DeNardo DG, Ruffell B. Macrophages as regulators of tumour immunity and immunotherapy. Nat Rev Immunol 2019;19:369-82. [Crossref] [PubMed]
- Hilligan KL, Ronchese F. Antigen presentation by dendritic cells and their instruction of CD4+ T helper cell responses. Cell Mol Immunol 2020;17:587-99. [Crossref] [PubMed]
- Liu S, Galat V, Galat Y, et al. NK cell-based cancer immunotherapy: from basic biology to clinical development. J Hematol Oncol 2021;14:7. [Crossref] [PubMed]
- Cristescu R, Mogg R, Ayers M, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science. 2018;362:eaar3593. Erratum in: Science 2019;363. [Crossref] [PubMed]
- Rizvi NA, Cho BC, Reinmuth N, et al. Durvalumab With or Without Tremelimumab vs Standard Chemotherapy in First-line Treatment of Metastatic Non-Small Cell Lung Cancer: The MYSTIC Phase 3 Randomized Clinical Trial. JAMA Oncol 2020;6:661-74. [Crossref] [PubMed]
- Mok TSK, Wu YL, Kudaba I, et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet 2019;393:1819-30. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Rooney MS, Shukla SA, Wu CJ, et al. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015;160:48-61. [Crossref] [PubMed]
- Liu D, Schilling B, Liu D, et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat Med 2019;25:1916-27. [Crossref] [PubMed]
- Hellmann MD, Nathanson T, Rizvi H, et al. Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer. Cancer Cell 2018;33:843-852.e4. [Crossref] [PubMed]
- Schrock A, Sharma N, Peled N, et al. MA14.01 Updated Data set Assessing Tumor Mutation Burden (TMB) as a Biomarker for Response to PD-1/PD-L1 Targeted Therapies in Lung Cancer (LC). J Thorac Oncol 2017;12:S422. [Crossref]
- Xiao Y, Freeman GJ. The microsatellite instable subset of colorectal cancer is a particularly good candidate for checkpoint blockade immunotherapy. Cancer Discov 2015;5:16-8. [Crossref] [PubMed]
- Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509-20. [Crossref] [PubMed]
(English Language Editor: J. Gray)


