Efficacy of artesunate in asthma: based on network pharmacology and molecular docking
Highlight box
Key findings
• Artesunate has the potential to be a potent and safe anti-asthmatic agent through diverse therapeutic mechanisms and acceptable safety.
What is known and what is new?
• Artesunate may be able to treat asthma through reversing airway hypersensitivity and glucocorticoid insensitivity, ameliorating inflammation, preventing mast cell degranulation, and inducing eosinophil death.
• In this study, we analyze the safety of artesunate in clinical application in asthma, and predict additional new mechanisms, such as prevent the occurrence of asthma by regulating the internal environment, the response to external stimuli, and the generation and survival of leukocytes, and improving the outcome of asthma by regulating the biosynthesis and metabolism of steroid hormone and reversing airway remodeling.
What is the implication, and what should change now?
• Asthmatic patients could benefit from artesunate for its high cost-effectiveness.
Introduction
Bronchial asthma is a common chronic respiratory disorders affecting approximately 358 million people around the world (1), characterized by variable presentations of wheeze, shortness of breath, cough, and chest tightness, in correlation with chronic airway inflammation, reversible expiratory airflow limitation, and airway hyperresponsiveness (AHR) (2). Severe asthma occurred in more than 10% of adults and 2.5% of children with asthma, causing impaired quality of life and increased risk of persistent airflow limitation, exacerbations, hospitalization and death (3), leading to intensified treatment such as an additional dose, frequency or duration of inhaled glucocorticoids, maintenance oral glucocorticoid, or biologic therapies, etc. (2,4,5). All the intensified therapies may bring about various adverse events and heavy economic burdens, such as infection (e.g., herpes zoster and parasites), neoplasm, obesity or metabolic disorders, asthma worsening, nasopharyngitis, etc. (5-8). Therefore, a cost-effective and safe drug with multiple targets is needed for anti-asthmatic treatment to comprehensively control and relieve asthma and, meanwhile, to alleviate the economic burdens of patients.
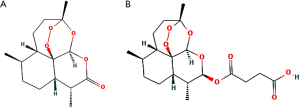
Artesunate, a semi-synthetic derivative of artemisinin which is isolated from Artemisia Annua L., a traditional Chinese herb, has been the first-line treatment against plasmodium falciparum malaria for decades (Figure 1A), which turns into dihydroartemisinin (DHA), the active metabolite, in a short time in vivo (Figure 1B). Recently, multiple experiments have been reported in the potential therapeutical effects of artesunate in asthma, including reversing airway hypersensitivity (9,10) and glucocorticoid insensitivity (11), ameliorating inflammation (12), preventing mast cell degranulation (13), and inducing eosinophil death (14-16), etc. However, there still remains a lack of comprehensive analysis and evaluation of artesunate and DHA in potential anti-asthmatic pharmacological mechanisms and toxicology in human studies.
In this study, we explore the potential mechanisms and evaluate the safety of artesunate and its metabolite, DHA, in asthma based on network pharmacology and bioinformatic analysis. We present the following article in accordance with the STROBE reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1437/rc).
Methods
This study was designed based on evaluating properties of artesunate and DHA, identifying targets of both compounds and asthma, screening out the overlapping targets, establishing a compound-target network and protein-protein interaction network, and conducting enrichment analysis and molecular docking. The treatment of artesunate and its metabolite in asthma was regarded effective, if there were effective interactions founded between hub targets and compounds.
Property evaluation
SMILES of artesunate and DHA were obtained from PubChem. Propertie of both compounds were evaluated on the SwissADME (http://www.swissadme.ch) (17,18) and ADMETlab 2.0 (https://admetmesh.scbdd.com) (19), including properties like physicochemistry properties, absorption, distribution, metabolism, excretion and toxicology to evaluate their druglikeness and safety. SwissADME is the most commonly used platform to understand the ADME characteristics of chemicals, while the ADMETlab can supplement some information mainly in toxicology such as toxicity, toxicophore rules, and more medicinal chemistry rules with a faster computation time (19). Combined materials from both platforms can provide a more comprehensive evaluations for both study molecules.
Targets of artesunate and its metabolite
Targets of artesunate and DHA were predicted from SwissTargetPrediction (http://swisstargetprediction.ch) (20,21) and PharmMapper (http://lilab-ecust.cn/pharmmapper/index.html) (22-24) by SMILES and 3-dimensional structure (both downloaded from PubChem), respectively. All potential targets were combined as a union set for more accurate results.
Targets of asthma
Targets of asthma were acquired from GeneCards (https://www.genecards.org) (25) and DisGeNET (https://disgenet.org) (26) databases by the term of ‘asthma’. All targets were combined as a union set for further analysis.
Compound-target network
To identify the relationship between common targets and compounds, compound-target network was constructed. Common targets of compounds and asthma were obtained by overlapping the potential target set of artesunate and DHA and the target set of asthma and visualized by the Venndiagram package in R version 4.1.2. Corresponding compound-target network was constructed by Cytoscape 3.8.0 where larger symbols were used to represent the common targets of both artesunate and DHA (27).
Protein-protein interaction network
To explore the interaction between the common genes, protein-protein interaction (PPI) network was established by String (https://cn.string-db.org) with the confidence of 0.4 and visualized in Cytoscape 3.8.0, where targets were rearranged with degree and displayed with gradient color and size based on the degree. The Maximal Clique Centrality (MCC) algorithm, which was regarded as the most effective method to find hub nodes, was used to identify the top 10 hub genes by cytoHubba, a plugin in Cytoscape 3.8.0 (27).
Enrichment analysis
Enrichment analyses were conducted on Metascape (https://metascape.org), including Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, Reactome Gene Set pathway, Gene Ontology (GO) Biological Process (GOBP), GO Molecular Function (GOMF), and GO Cellular Component (GOCC), with a minimal overlap of 3, a P value cutoff of 0.01, and minimal enrichment of 1.5 (28). Corresponding bubble plots were made in R version 4.1.2 where the bubble size represented the gene count and the color represented −lgP.
Molecular docking
Ten hub genes were identified as mentioned above and used for further molecular docking to investigate the interaction between the drugs and the targets. First, three-dimensional structures of hub proteins without or with a ligand similar to the study compounds were obtained from RCSB Protein Data Bank (PDB, https://www.rcsb.org) (29-31). Structures of artesunate and DHA were downloaded from Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP, https://old.tcmsp-e.com/index.php) (32). Second, through AutoDock Tools 1.5.7, water was deleted and all hydrogens were added for proteins, and all hydrogens were added and charge balance and rotatable bonds were detected for each molecule. Third, the whole protein or, at least, the receptor active center was enclosed in a grid for further dockings. Forth, the semi-flexible dockings of receptors and corresponding ligands were performed by AutoDock Vina with default settings. Then the optimal docking structures were output based on the over-all best vina energy (less than −4 kcal/mol, based on the rule of thumb), hydrogen bonds and active pockets of original ligands to select the most stable receptor-ligand complex. Finally, the results were visualized including interacting residues and hydrogen bonds in the PyMOL Molecular Graphics System (Version 2.5.2, Schrödinger, LLC).
Results
Property evaluation of Artesunate and DHA
Canonic SMILES of artesunate [CID: 6917864; CC1CCC2C(C(OC3C24C1CCC(O3)(OO4)C)OC(=O)CCC(=O)O)C) and DHA (CID: 3000518; CC1CCC2C(C(OC3C24C1CCC(O3)(OO4)C)O)C] were acquired from PubChem.
Properties of artesunate and DHA obtained from SwissADME and ADMETlab indicated that both compounds had desirable performances for clinical application (Figure 2 and Table 1). Two molecules are both suitable to be drugs according to various druglikeness evaluation criteria, such as Lipinski rules, Ghose rules, Veber rules, Egan rules, Muegge rules, Bioavailability Score, Pfizer rules, GSK rules, GoldenTriangle rules, etc. High GI absorption indicated their desirable oral bioavailability. Safety and frequent administration was suggested based on the high clearance and relatively short half-life. Toxicology evaluations showed adverse events that needed special attention in further clinical practice mainly involved hepatotoxicity (H-HT and DILI), lung injury (respiratory toxicology), mutagenicity (AMES toxicity) and carcinogenicity (Table 1).
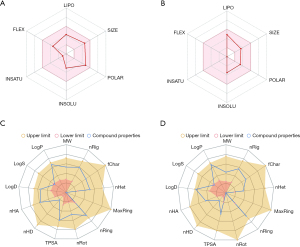
Table 1
| Properties | Indicator | Artesunate | DHA |
|---|---|---|---|
| Physicochemistry properties | MW | 384.42 | 284.35 |
| Rotatable bonds | 5 | 0 | |
| H-bond acceptors | 8 | 5 | |
| H-bond donors | 1 | 1 | |
| TPSA | 100.52 | 57.15 | |
| Consensus Log Po/w | 2.07 | 2.25 | |
| Water Solubility | Soluble | Soluble | |
| Druglikeness | Lipinski violations | 0 | 0 |
| Ghose violations | 0 | 0 | |
| Veber violations | 0 | 0 | |
| Egan violations | 0 | 0 | |
| Muegge violations | 0 | 0 | |
| Bioavailability score | 0.56 | 0.55 | |
| Pfizer | Accepted | Accepted | |
| GSK | Accepted | Accepted | |
| GoldenTriangle | Accepted | Accepted | |
| Absorption | GI absorption | High | High |
| Pgp-substrate | No | No | |
| Distribution | BBB permeant | No | Yes |
| Log Kp (skin permeation, cm/s) | −7.31 | −5.91 | |
| PPB | 60.43% | 85.44% | |
| Metabolism | CYP1A2 inhibitor | No | Yes |
| CYP2C19 inhibitor | No | No | |
| Metabolism | CYP2C9 inhibitor | No | No |
| CYP2D6 inhibitor | No | No | |
| CYP3A4 inhibitor | No | No | |
| Excretion | CL (mL/min/kg) | 14.450 | 15.838 |
| T1/2 score | 0.549 | 0.181 | |
| Toxicology | PAINS alerts | 0 | 0 |
| Brenk alerts | Peroxide | Peroxide | |
| Leadlikeness violations | MW>350 | 0 | |
| Synthetic accessibility | 6.67 | 6.59 | |
| Toxicophores | Peroxide | Peroxide | |
| SureChEMBL | Peroxide | Peroxide | |
| Nongenotoxic carcinogenicity | 0 | 0 | |
| Genotoxic carcinogenicity rule | 0 | 0 | |
| hERG blockers | −−− | − | |
| H-HT | +++ | +++ | |
| DILI | ++ | − | |
| AMES toxicity | +++ | ++ | |
| FDAMDD | −−− | −− | |
| Skin sensitivity | − | −− | |
| Carcinogenicity | + | ++ | |
| Eye corrosion | −−− | −−− | |
| Eye irritation | −−− | −−− | |
| Respiratory toxicology | ++ | +++ |
DHA, dihydroartemisinin; MW, molecular weight; TPSA, topological polar surface area; GSK, GlaxoSmithKline; GI, gastrointestinal; BBB, blood-brain barrier; PPB, plasma protein binding; H-HT, human hepatotoxicity; DILI, drug-induced liver injury; AMES, Ames test for mutagenicity; FDAMDD, Food and Drug Administration maximum recommended daily dose.
Targets of artesunate and DHA
For artesunate, 86 target genes were obtained from SwissTargetPrediction, and 67 from PharmMapper, while for DHA, 97 potential targets were acquired from SwissTargetPrediction, and 67 from PharmMapper. After removing the duplicated data, we got a union set of 282 targets.
Targets of asthma
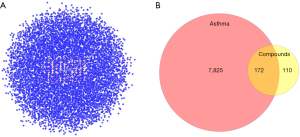
When searching for the potential therapeutical targets of asthma, we obtained 7,490 targets from GeneCards, and 2,096 targets from DisGeNET. A total of 7,997 targets were acquired based on both databases.
Compound-target network
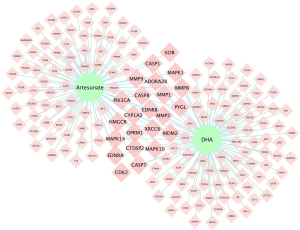
A total of 172 overlapping targets were identified between the molecules and asthma (Figure 3). A compound-target network was established with 23 common target genes between artesunate and DHA (Figure 4), including CYP1A2, HMGCR, EDNRB, EDNRA, CASP1, CASP7, MAPK1, CASP8, PYGL, MDM2, MAPK14, MAPK10, MMP1, MMP2, PIK3CA, OPRM1, KDR, CDK2, MMP9, MMP8, ADORA2B, CTDSP2, and XRCC6.
Protein-protein interaction network and Hub genes
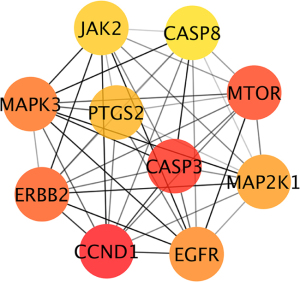
A protein-protein interaction (PPI) network was constructed with the total 172 common targets and visualized in Cytoscape. Genes were rearranged with gradient sizes and colors based on the degrees (Figure 5). After calculating with MCC algorithm, we got 10 hub genes, namely CCND1, CASP3, MTOR, ERBB2, MAPK3, EGFR, MAP2K1, PTGS2, JAK2, CASP8 (see Table 2 and Figure 6).
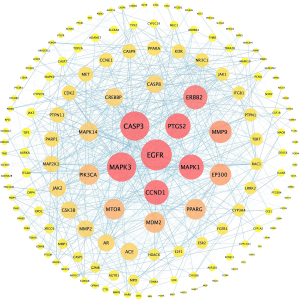
Table 2
| Rank | Targets | Score | Corresponding compounds |
|---|---|---|---|
| 1 | CCND1 | 9.72E+10 | Artesunate |
| 2 | CASP3 | 9.71E+10 | Artesunate |
| 3 | MTOR | 9.71E+10 | DHA |
| 4 | ERBB2 | 9.66E+10 | DHA |
| 5 | MAPK3 | 9.66E+10 | DHA |
| 6 | EGFR | 9.65E+10 | DHA |
| 7 | MAP2K1 | 9.64E+10 | DHA |
| 8 | PTGS2 | 9.63E+10 | DHA |
| 9 | JAK2 | 9.56E+10 | DHA |
| 10 | CASP8 | 9.56E+10 | Both |
DHA, dihydroartemisinin.
Biofunction enrichment analysis
In order to investigate the potential therapeutic mechanisms that artesunate and DHA may be involved in, we conducted different enrichment analyses to explore the possible pathological and physiological processes, molecular functions and target cell components including KEGG pathway, Reactome Gene Sets, GO biological process, GO molecular function and GO cell component.
Top 20 results were ranked in Figure 7. In Figure 7A-7C, artesunate and its metabolite were found participating in various pathways of the development and improvement of asthma. The investigated molecules may synergize the therapeutic effects of glucocortoid by enhancing the glucocorticoid sensitivity through regulating the biosynthesis and metabolism of and response to steroids. Meanwhile, they might play a role in alleviating the airway hyperresponsiveness (AHR) through regulating the response to multiple internal and external stimuli, such as stress, hormone, lipopolysaccharide (LPS), inorganic substance, decreased oxygen levels, etc. What’s more, these two compounds may modulate immunity and inflammation responses through interleukin signaling, neuroactive ligand-receptor interaction, NF-kappa B signaling pathway, Fc epsilon receptor (FCERI) signaling, MAPK3 (ERK1) activation, Toll-like receptor cascades, hematopoietic or lymphoid organ development, serotonergic synapse, etc. In addition, the airway remodeling in the development of chronic asthma may be reversed by artesunate (and DHA) through affecting the gland development and extracellular matrix organization. Furthermore, diverse regulations in cell survival and death may affect multiple participant cells in asthma, such as increased and activated eosinophils and proliferative smooth muscle cells.
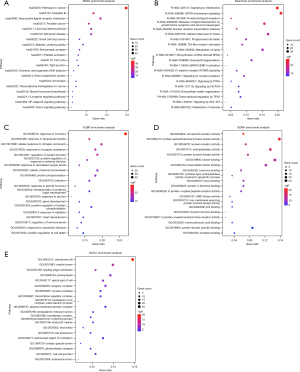
Multiple molecular functions were found by GO analysis, such as eicosanoid receptor activity, protein serine/threonine/tyrosine kinase activity, nuclear receptor activity, etc., which were associated with steroid hormone receptor activity, cell survival and death, and inflammation response, etc. (Figure 7D). Main cell components the overlapping genes cluster included membrane raft, vesicle lumen, and leading edge membrane (Figure 7E).
Molecular docking
Further, we performed molecular docking to figure out the interaction between the small molecules (artesunate and DHA) and hub proteins so as to screen most potential targets and corresponding binding sites. PDB database was used to acquire hub target proteins without or with ligands of similar structures to corresponding compounds, that is, G1/S-specific cyclin-D1 (gene symbol: CCND1; PDB ID: 6P8E) (33), Caspase-3 (gene symbol: CASP3; PDB ID: 1RHU) (34), mTOR (gene symbol: MTOR; PDB ID: 4JT5) (35), ErbB2 (gene symbol: ERBB2; PDB ID: 3PP0) (36), Mitogen-activated protein kinase (MAPK3; gene symbol: MAPK3; PDB ID: 2ZOQ) (37), epidermal growth factor receptor (EGFR; gene symbol: EGFR; PDB ID: 1M17) (38), dual specificity mitogen-activated protein kinase kinase 1 (gene symbol: MAP2K1; PDB ID: 3ZLW) (39), Prostaglandin G/H synthase 2 (PGHS-2; gene symbol: PTGS2; PDB ID: 5KIR) (40), Janus kinase 2 (JAK-2; gene symbol: JAK2; PDB ID: 3KRR) (41), and Caspase-8 (gene symbol: CASP8; PDB ID: 4ZBW) (42). Structures of artesunate (MOL007434) and DHA (MOL007425) were obtained by TCMSP.
Through AutoDock Vina, 10 optimal stable interactions between 9 targets (except caspase-3) and corresponding compounds were identified based on the vina binding energy value, hydrogen bond formation and confirmed active pockets for original ligands (Figure 8). Corresponding binding energy values were presented and ranked by binding energy in Table 3, namely PGHS-2-DHA (−8.4 kcal/mol), MAPK3-DHA (−7.6 kcal/mol), JAK2-DHA (−7.6 kcal/mol), mTOR-DHA (−7.3 kcal/mol), CASP8-Artesunate (−7.3 kcal/mol), CASP8-DHA (−6.5 kcal/mol), ErbB2-DHA (−6.3 kcal/mol), MAP2K1-DHA (−5.9 kcal/mol), CCND1-Artesunate (−5.8 kcal/mol), and EGFR-DHA (−4.7 kcal/mol). All the 10 dockings indicated relatively stable interactions between the receptor and the corresponding ligands.
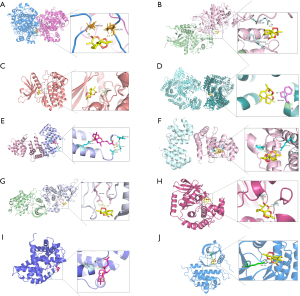
Table 3
| Rank | Targets | Corresponding compounds | Binding energy (kcal/mol) |
|---|---|---|---|
| 1 | PGHS-2 | DHA | −8 |
| 2 | MAPK3 | DHA | −7.6 |
| 3 | JAK2 | DHA | −7.6 |
| 4 | mTOR | DHA | −7.3 |
| 5 | CASP8 | Artesunate | −7.3 |
| 6 | CASP8 | DHA | −6.5 |
| 7 | ErbB2 | DHA | −6.3 |
| 8 | MAP2K1 | DHA | −5.9 |
| 9 | CCND1 | Artesunate | −5.8 |
| 10 | EGFR | DHA | −4.7 |
| 11 | CASP3 | Artesunate | 0 |
DHA, dihydroartemisinin.
Discussion
Druglikeness and safety evaluation
In this study, we made a preliminary evaluation of artesunate (as well as its active metabolite, DHA) in therapeutic mechanisms and safety in treating asthma. Properties in physicochemistry, absorption, distribution, metabolism, excretion and toxicology showed that artesunate is a safe candidate for respiratory disorders with desirable druglikeness, acceptable oral bioavailability, quick metabolism and excretion, and relatively low toxicity. As for the alerts about the respiratory toxicity, previous studies have shown that artesunate showed no cytotoxicity on healthy, non-diseased cells, including human normal lung epithelial cells (BEAS-2B) (43), normal human lung fibroblast (WI-38) (44,45), non-cancerous human dermal fibroblasts (CCD-1108Sk) (46), and normal hepatic cells (L-02) (47), providing more reliable evidence on its safety in clinical application.
Biofunctional prediction
Further, we explored the potential therapeutic mechanisms of artesunate based on network pharmacology, a powerful method to obtain a systemic understanding of chemicals. In this study, actually, we searched SwissTargetPrediction, PharmMapper, Drugbank (https://go.drugbank.com) and TCMSP databases for the potential targets, but no targets of homo sapiens were found in the latter two platforms. Thus, 282 targets of artesunate and DHA were acquired. Besides, 7,997 genes were obtained that play a role in bronchial asthma from GeneCards and DisGeNET. Finally, 172 intersected targets were identified by overlapping these two gene sets.
According to the biofunction analysis, artesunate and DHA showed great potential in preventing, controlling and relieving bronchial asthma through multiple pathophysiologic and therapeutic processes, including biosynthesis of, metabolism of and response to steroid hormone, immune and inflammatory response, airway hyperresponsiveness, airway remodeling and cell survival and death regulation. Results showed that artesunate and its metabolite may prevent the occurence of asthma by regulating the internal environment through immune and inflammatory pathways, the response to external stimuli, and the generation and survival of leukocytes. Further experiments are needed to figure out the prevention effects of artesunate on asthma. Moreover, such agents may relieve the acute exacerbations of asthma by reversing the airway hyperresponsiveness and inflammation. An animal experiment have proved that artesunate could decrease the airway resistance and the contraction of airway smooth cells by lowering the intracellular calcium ion (Ca2+) concentration through binding with G protein coupled bitter taste receptors (TAS2Rs) in the ASMCs (9). More in vivo and in vitro experiments are needed on the effects of the potential targets identified in this in-silico study. Additionally, during the conventional treatment of asthma, particularly difficult-to-treat asthma, artesunate was predicted to play a special role in modulating biosynthesis of, metabolism of and response to glucocorticoid so as to enhance the therapeutic effects and reduce the severe adverse effects of high-dose and long-term glucocorticoid. Our research team has found that artesunate could reverse CSE-induced glucocorticoid insensitivity and restored HDAC2 deactivation induced by CSE (11). What’s more, considering that cell cycle arrest and cell death induction are the main clustering functions in this study and, meanwhile, have been proved before in multiple oncologic studies, including apoptosis, autophagy, ferroptosis, etc. (44,46,48-53), combined with the development of asthma, we deduced that artesunate and its metabolite could alleviating asthma by decreasing the eosinophil count and smooth muscle count in airways. Relative studies performed by our team and others have partially proved the hypothesis (14-16), but complete explanations in detail are still needed. Besides, artesunate may function in suppressing the chronic progression of asthma by inhibiting airway remodeling through affecting extracellular matrix organization and smooth cell growth and proliferation. In an in-vitro experiment on primary human cultured airway smooth muscle cells (ASMC), proliferation of ASMCs and subsequent inflammation and oxidative stress was inhibited by artesunate through PI3K/Akt/p70S6K and p42/p44 mitogen-activated protein kinases (MAPK) pathways (10), supporting our in-silico predictions. On the other hand, previous studies have also found that artesunate and DHA could prevent mast cells from degranulation (13), indicating that artesunate may treat asthma through much more pathways than we predicted in this study. From the other perspective, GO molecular function and cell component analysis provide additional information about the target locations and functions in a molecular level for further wet-lab experiments.
Receptor-ligand interaction analysis
Among the 11 topological analysis methods in the Cytoscape plugin, cytoHubba, MCC performs better than the others, which captures more essential proteins in the top ranked list in both high-degree and low-degree proteins (54). Therefore, this study used MCC to identify the hub genes. Combined with the molecular docking results, 9 targets were screened out according to the binding energy, hydrogen bonds, and active pockets of the original ligands, including PGHS-2, MAPK3, JAK2, mTOR, CASP8, ErbB2, MAP2K1, CCND1, and EGFR, in rank order. Among these interactions, those with binding energy values lower than −6 kcal/mol could be regarded as strong affinity with biologic activity based on the rule of thumb and relative analysis (55), including PGHS-2, MAPK3, JAK2, and mTOR with DHA, CASP8 with artesunate and DHA, and ErbB2 with DHA. Among these, prostaglandin G/H synthase 2, also known as cyclooxygenase-2 (COX-2), always plays an essential role in inflammatory response (56,57) and take the primary responsibility for the prostaglandin production in immune cells (58). In asthmatic mouse models and patients, PTGS2 and its catalyzed product, PGE2, were found being up-regulated and associated with allergic inflammation (59-63). What’s more, previous studies have also indicated that PTGS2 regulated the activity of Th1, Th2, and Th17 cells (61,64,65), and PGE2 was associated with exacerbations of allergen-induced pulmonary inflammation (66), systemic inflammation (67), and IgE production (68) in vivo. MAPK3/ERK2 takes a significant part in the MAPK/ERK cascade, regulating diverse intercellular activities, such as transcription, translation, cytoskeletal rearrangement, so as to mediate cell growth, adhesion, survival and differentiation, etc., especially after stimuli (69-72). Studies suggested that MAPK3/ERK2 pathway mainly participates in airway remodeling during the development of asthma, including lung fibroblast (73) and ASMCs proliferation (74,75). What’s more, activation of ERK1 and ERK2 pathways in eosinophils stimulated by IL-5 contributes to synthesis of leukotriene C4 in the eosinophils (76). Besides, these signaling pathways can induce neutrophil recruitment by ASMCs leading to insensitivity to glucocorticoids in difficult-to-control asthma (77). JAK2 was expressed most in the spleen and peripheral blood leukocytes, especially eosinophils (78). Activated by IL-5, JAK2 could promote eosinophil survival through anti-apoptotic effects (79-83). According to a clinical case series, severe asthma with blood hypereosinophilia was proved to be associated with JAK2 V617F mutations (84). Serine/threonine-protein kinase mTOR was activated at the onset of asthma and suppressed during the recovery, and suppressing the mTOR pathway in asthmatic mice could inhibit subsequent inflammatory processes and normalize the balance of Th17/Treg and Th1/Th2 cytokines (85,86). For caspase-8, one of the chromosomal regions contributing to the development of asthma and allergic disorders was found including caspase-8, and further tests on its single nucleotide polymorphisms (SNPs) indicated an association between caspase-8 and the severity of AHR (as determined by PC20) in race-specific analysis (87). Combined with its main functions in programmed cell death (88-91), hypothesis could be made that artesunate may alleviate AHR through inducing the programmed cell death of ASMCs. Further investigations are needed. ErbB-2, an epithelial growth factor (EGF) family receptor, was proved to be a barrier to normal activity and repair of airway epithelial cell repair in asthma (92-95). Multiple previous studies on the pathophysiological processes and therapeutic targets of asthma provided theoretical evidence for artesunate to treat asthma from various pathways, but, still, there need experiments and clinical trials to confirm them.
There are limitations in this study. First, although we made a comprehensive evaluation and analysis based on various platforms and databases from multiple aspects, the conclusions are, from source, based on in-silico analysis. Considering the inherent limitations of computational analysis, web-lab experiments should be conducted further to clarify the real mechanisms. Second, asthma is a heterogeneous disease with diverse clinical presentations, types, and severity, but we only provided a systemic analysis based on bronchial asthma ignoring its subtypes. According to the results, further analysis can be performed based on difficult-to-control asthma, severe asthma, allergic asthma, exacerbations of asthma, and chronic asthma, etc. But no matter which target sets are utilized for analysis, the results still need to be confirmed by web-lab experiments and clinical trials. Therefore, the potential mechanism of artesunate still needs more experiments to explore.
Conclusions
Artesunate has the potential to be a potent and safe anti-asthmatic agent based on its diverse therapeutic mechanisms and acceptable safety in silico.
Acknowledgments
Funding: This study was supported by the National Natural Science Foundation of China (Grant No. 82170027).
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1437/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1437/dss
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-22-1437/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Global, regional, and national deaths, prevalence, disability-adjusted life years, and years lived with disability for chronic obstructive pulmonary disease and asthma, 1990-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Respir Med 2017;5:691-706. [Crossref] [PubMed]
- Global Initiative for Asthma. Global Strategy for Asthma Management and Prevention, 2021. Available online: https://ginasthma.org/reports
- Settipane RA, Kreindler JL, Chung Y, et al. Evaluating direct costs and productivity losses of patients with asthma receiving GINA 4/5 therapy in the United States. Ann Allergy Asthma Immunol 2019;123:564-572.e3. [Crossref] [PubMed]
- Szefler SJ, Martin RJ, King TS, et al. Significant variability in response to inhaled corticosteroids for persistent asthma. J Allergy Clin Immunol 2002;109:410-8. [Crossref] [PubMed]
- Barnes PJ. Glucocorticoids and Asthma. In: Encyclopedia of Hormones. Academic Press; 2003:127-134.
- Khatri S, Moore W, Gibson PG, et al. Assessment of the long-term safety of mepolizumab and durability of clinical response in patients with severe eosinophilic asthma. J Allergy Clin Immunol 2019;143:1742-1751.e7. [Crossref] [PubMed]
- Ortega HG, Liu MC, Pavord ID, et al. Mepolizumab treatment in patients with severe eosinophilic asthma. N Engl J Med 2014;371:1198-207. [Crossref] [PubMed]
- FitzGerald JM, Bleecker ER, Nair P, et al. Benralizumab, an anti-interleukin-5 receptor α monoclonal antibody, as add-on treatment for patients with severe, uncontrolled, eosinophilic asthma (CALIMA): a randomised, double-blind, placebo-controlled phase 3 trial. Lancet 2016;388:2128-41. [Crossref] [PubMed]
- Wang Y, Wang A, Zhang M, et al. Artesunate attenuates airway resistance in vivo and relaxes airway smooth muscle cells in vitro via bitter taste receptor-dependent calcium signalling. Exp Physiol 2019;104:231-43. [PubMed]
- Tan SS, Ong B, Cheng C, et al. The antimalarial drug artesunate inhibits primary human cultured airway smooth muscle cell proliferation. Am J Respir Cell Mol Biol 2014;50:451-8. [Crossref] [PubMed]
- Luo Q, Lin J, Zhang L, et al. The anti-malaria drug artesunate inhibits cigarette smoke and ovalbumin concurrent exposure-induced airway inflammation and might reverse glucocorticoid insensitivity. Int Immunopharmacol 2015;29:235-45. [Crossref] [PubMed]
- Cheng C, Ho WE, Goh FY, et al. Anti-malarial drug artesunate attenuates experimental allergic asthma via inhibition of the phosphoinositide 3-kinase/Akt pathway. PLoS One 2011;6:e20932. [Crossref] [PubMed]
- Cheng C, Ng DS, Chan TK, et al. Anti-allergic action of anti-malarial drug artesunate in experimental mast cell-mediated anaphylactic models. Allergy 2013;68:195-203. [Crossref] [PubMed]
- Wu Y, Chen H, Xuan N, et al. Induction of ferroptosis-like cell death of eosinophils exerts synergistic effects with glucocorticoids in allergic airway inflammation. Thorax 2020;75:918-27. [Crossref] [PubMed]
- Wang R, Lin J, Wang J, Li C. Effects of artesunate on eosinophil apoptosis and expressions of Fas and Bcl-2 proteins in asthmatic mice. Nan Fang Yi Ke Da Xue Xue Bao 2020;40:93-8. [PubMed]
- Wang RY, Li HW, Zhang Q, Lin JT. Effect of artesunate on airway responsiveness and airway inflammation in asthmatic mice. Zhonghua Yi Xue Za Zhi 2019;99:2536-41. [PubMed]
- Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 2017;7:42717. [Crossref] [PubMed]
- Daina A, Michielin O, Zoete V. iLOGP: a simple, robust, and efficient description of n-octanol/water partition coefficient for drug design using the GB/SA approach. J Chem Inf Model 2014;54:3284-301. [Crossref] [PubMed]
- Xiong G, Wu Z, Yi J, et al. ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res 2021;49:W5-W14. [Crossref] [PubMed]
- Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res 2019;47:W357-64. [Crossref] [PubMed]
- Gfeller D, Michielin O, Zoete V. Shaping the interaction landscape of bioactive molecules. Bioinformatics 2013;29:3073-9. [Crossref] [PubMed]
- Liu X, Ouyang S, Yu B, et al. PharmMapper server: a web server for potential drug target identification using pharmacophore mapping approach. Nucleic Acids Res 2010;38:W609-14. [Crossref] [PubMed]
- Wang X, Shen Y, Wang S, et al. PharmMapper 2017 update: a web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res 2017;45:W356-60. [Crossref] [PubMed]
- Wang X, Pan C, Gong J, et al. Enhancing the Enrichment of Pharmacophore-Based Target Prediction for the Polypharmacological Profiles of Drugs. J Chem Inf Model 2016;56:1175-83. [Crossref] [PubMed]
- Safran M, Rosen N, Twik M, et al. The GeneCards Suite. In: Abugessaisa I, Kasukawa T, eds. Practical Guide to Life Science Databases. Springer Singapore; 2021:27-56.
- Piñero J, Ramírez-Anguita JM, Saüch-Pitarch J, et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res 2020;48:D845-55. [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10:1523. [Crossref] [PubMed]
- Burley SK, Bhikadiya C, Bi C, et al. RCSB Protein Data Bank: powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res 2021;49:D437-51. [Crossref] [PubMed]
- Berman HM, Westbrook J, Feng Z, et al. The Protein Data Bank. Nucleic Acids Res 2000;28:235-42. [Crossref] [PubMed]
- Berman H, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat Struct Biol 2003;10:980. [Crossref] [PubMed]
- Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform 2014;6:13. [Crossref] [PubMed]
- Guiley KZ, Stevenson JW, Lou K, et al. p27 allosterically activates cyclin-dependent kinase 4 and antagonizes palbociclib inhibition. Science 2019;366:eaaw2106. [Crossref] [PubMed]
- Becker JW, Rotonda J, Soisson SM, et al. Reducing the peptidyl features of caspase-3 inhibitors: a structural analysis. J Med Chem 2004;47:2466-74. [Crossref] [PubMed]
- Yang H, Rudge DG, Koos JD, et al. mTOR kinase structure, mechanism and regulation. Nature 2013;497:217-23. [Crossref] [PubMed]
- Aertgeerts K, Skene R, Yano J, et al. Structural analysis of the mechanism of inhibition and allosteric activation of the kinase domain of HER2 protein. J Biol Chem 2011;286:18756-65. [Crossref] [PubMed]
- Kinoshita T, Yoshida I, Nakae S, et al. Crystal structure of human mono-phosphorylated ERK1 at Tyr204. Biochem Biophys Res Commun 2008;377:1123-7. [Crossref] [PubMed]
- Stamos J, Sliwkowski MX, Eigenbrot C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J Biol Chem 2002;277:46265-72. [Crossref] [PubMed]
- Amaning K, Lowinski M, Vallee F, et al. The use of virtual screening and differential scanning fluorimetry for the rapid identification of fragments active against MEK1. Bioorg Med Chem Lett 2013;23:3620-6. [Crossref] [PubMed]
- Orlando BJ, Malkowski MG. Crystal structure of rofecoxib bound to human cyclooxygenase-2. Acta Crystallogr F Struct Biol Commun 2016;72:772-6. [Crossref] [PubMed]
- Baffert F, Régnier CH, De Pover A, et al. Potent and selective inhibition of polycythemia by the quinoxaline JAK2 inhibitor NVP-BSK805. Mol Cancer Ther 2010;9:1945-55. [Crossref] [PubMed]
- Shen C, Yue H, Pei J, et al. Crystal structure of the death effector domains of caspase-8. Biochem Biophys Res Commun 2015;463:297-302. [Crossref] [PubMed]
- Ravindra KC, Ho WE, Cheng C, et al. Untargeted Proteomics and Systems-Based Mechanistic Investigation of Artesunate in Human Bronchial Epithelial Cells. Chem Res Toxicol 2015;28:1903-13. [Crossref] [PubMed]
- Ganguli A, Choudhury D, Datta S, et al. Inhibition of autophagy by chloroquine potentiates synergistically anti-cancer property of artemisinin by promoting ROS dependent apoptosis. Biochimie 2014;107 Pt B:338-49.
- Mi YJ, Geng GJ, Zou ZZ, et al. Dihydroartemisinin inhibits glucose uptake and cooperates with glycolysis inhibitor to induce apoptosis in non-small cell lung carcinoma cells. PLoS One 2015;10:e0120426. [Crossref] [PubMed]
- Rassias DJ, Weathers PJ. Dried leaf Artemisia annua efficacy against non-small cell lung cancer. Phytomedicine 2019;52:247-53. [Crossref] [PubMed]
- Xu CC, Deng T, Fan ML, et al. Synthesis and in vitro antitumor evaluation of dihydroartemisinin-cinnamic acid ester derivatives. Eur J Med Chem 2016;107:192-203. [Crossref] [PubMed]
- Tong Y, Liu Y, Zheng H, et al. Artemisinin and its derivatives can significantly inhibit lung tumorigenesis and tumor metastasis through Wnt/β-catenin signaling. Oncotarget 2016;7:31413-28. [Crossref] [PubMed]
- Zhang Q, Yi H, Yao H, et al. Artemisinin Derivatives Inhibit Non-small Cell Lung Cancer Cells Through Induction of ROS-dependent Apoptosis/Ferroptosis. J Cancer 2021;12:4075-85. [Crossref] [PubMed]
- Zhou C, Pan W, Wang XP, et al. Artesunate induces apoptosis via a Bak-mediated caspase-independent intrinsic pathway in human lung adenocarcinoma cells. J Cell Physiol 2012;227:3778-86. [Crossref] [PubMed]
- Zhao Y, Jiang W, Li B, et al. Artesunate enhances radiosensitivity of human non-small cell lung cancer A549 cells via increasing NO production to induce cell cycle arrest at G2/M phase. Int Immunopharmacol 2011;11:2039-46. [Crossref] [PubMed]
- Liu WM, Gravett AM, Dalgleish AG. The antimalarial agent artesunate possesses anticancer properties that can be enhanced by combination strategies. Int J Cancer 2011;128:1471-80. [Crossref] [PubMed]
- Cheong DHJ, Tan DWS, Wong FWS, et al. Anti-malarial drug, artemisinin and its derivatives for the treatment of respiratory diseases. Pharmacol Res 2020;158:104901. [Crossref] [PubMed]
- Chin CH, Chen SH, Wu HH, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014;8:S11. [Crossref] [PubMed]
- Shityakov S, Förster C. In silico predictive model to determine vector-mediated transport properties for the blood-brain barrier choline transporter. Adv Appl Bioinform Chem 2014;7:23-36. [Crossref] [PubMed]
- Appleton I, Tomlinson A, Willoughby DA. Induction of cyclo-oxygenase and nitric oxide synthase in inflammation. Adv Pharmacol 1996;35:27-78. [Crossref] [PubMed]
- Kim SF, Huri DA, Snyder SH. Inducible nitric oxide synthase binds, S-nitrosylates, and activates cyclooxygenase-2. Science 2005;310:1966-70. [Crossref] [PubMed]
- Williams CS, Mann M, DuBois RN. The role of cyclooxygenases in inflammation, cancer, and development. Oncogene 1999;18:7908-16. [Crossref] [PubMed]
- Herrerias A, Torres R, Serra M, et al. Activity of the cyclooxygenase 2-prostaglandin-E prostanoid receptor pathway in mice exposed to house dust mite aeroallergens, and impact of exogenous prostaglandin E2. J Inflamm (Lond) 2009;6:30. [Crossref] [PubMed]
- Sousa Ar, Pfister R, Christie PE, et al. Enhanced expression of cyclo-oxygenase isoenzyme 2 (COX-2) in asthmatic airways and its cellular distribution in aspirin-sensitive asthma. Thorax 1997;52:940-5. [Crossref] [PubMed]
- Rumzhum NN, Patel BS, Prabhala P, et al. IL-17A increases TNF-α-induced COX-2 protein stability and augments PGE2 secretion from airway smooth muscle cells: impact on β2 -adrenergic receptor desensitization. Allergy 2016;71:387-96. [Crossref] [PubMed]
- Deacon K, Knox AJ. Human airway smooth muscle cells secrete amphiregulin via bradykinin/COX-2/PGE2, inducing COX-2, CXCL8, and VEGF expression in airway epithelial cells. Am J Physiol Lung Cell Mol Physiol 2015;309:L237-49. [Crossref] [PubMed]
- Profita M, Sala A, Bonanno A, et al. Increased prostaglandin E2 concentrations and cyclooxygenase-2 expression in asthmatic subjects with sputum eosinophilia. J Allergy Clin Immunol 2003;112:709-16. [Crossref] [PubMed]
- Carey MA, Germolec DR, Bradbury JA, et al. Accentuated T helper type 2 airway response after allergen challenge in cyclooxygenase-1-/- but not cyclooxygenase-2-/- mice. Am J Respir Crit Care Med 2003;167:1509-15. [Crossref] [PubMed]
- Li H, Edin ML, Bradbury JA, et al. Cyclooxygenase-2 inhibits T helper cell type 9 differentiation during allergic lung inflammation via down-regulation of IL-17RB. Am J Respir Crit Care Med 2013;187:812-22. [Crossref] [PubMed]
- Church RJ, Jania LA, Koller BH. Prostaglandin E(2) produced by the lung augments the effector phase of allergic inflammation. J Immunol 2012;188:4093-102. [Crossref] [PubMed]
- Trebino CE, Stock JL, Gibbons CP, et al. Impaired inflammatory and pain responses in mice lacking an inducible prostaglandin E synthase. Proc Natl Acad Sci U S A 2003;100:9044-9. [Crossref] [PubMed]
- Gao Y, Zhao C, Wang W, et al. Prostaglandins E2 signal mediated by receptor subtype EP2 promotes IgE production in vivo and contributes to asthma development. Sci Rep 2016;6:20505. [Crossref] [PubMed]
- Lavoie H, Gagnon J, Therrien M. ERK signalling: a master regulator of cell behaviour, life and fate. Nat Rev Mol Cell Biol 2020;21:607-32. [Crossref] [PubMed]
- Boulton TG, Nye SH, Robbins DJ, et al. ERKs: a family of protein-serine/threonine kinases that are activated and tyrosine phosphorylated in response to insulin and NGF. Cell 1991;65:663-75. [Crossref] [PubMed]
- Boulton TG, Yancopoulos GD, Gregory JS, et al. An insulin-stimulated protein kinase similar to yeast kinases involved in cell cycle control. Science 1990;249:64-7. [Crossref] [PubMed]
- Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev 2011;75:50-83. [Crossref] [PubMed]
- Matthiesen S, Bahulayan A, Holz O, et al. MAPK pathway mediates muscarinic receptor-induced human lung fibroblast proliferation. Life Sci 2007;80:2259-62. [Crossref] [PubMed]
- Vichi P, Whelchel A, Knot H, et al. Endothelin-stimulated ERK activation in airway smooth-muscle cells requires calcium influx and Raf activation. Am J Respir Cell Mol Biol 1999;20:99-105. [Crossref] [PubMed]
- Lee JH, Johnson PR, Roth M, et al. ERK activation and mitogenesis in human airway smooth muscle cells. Am J Physiol Lung Cell Mol Physiol 2001;280:L1019-29. [Crossref] [PubMed]
- Bates ME, Green VL, Bertics PJ. ERK1 and ERK2 activation by chemotactic factors in human eosinophils is interleukin 5-dependent and contributes to leukotriene C(4) biosynthesis. J Biol Chem 2000;275:10968-75. [Crossref] [PubMed]
- Robins S, Roussel L, Schachter A, et al. Steroid-insensitive ERK1/2 activity drives CXCL8 synthesis and neutrophilia by airway smooth muscle. Am J Respir Cell Mol Biol 2011;45:984-90. [Crossref] [PubMed]
- Saltzman A, Stone M, Franks C, et al. Cloning and characterization of human Jak-2 kinase: high mRNA expression in immune cells and muscle tissue. Biochem Biophys Res Commun 1998;246:627-33. [Crossref] [PubMed]
- Simon HU, Yousefi S, Dibbert B, et al. Anti-apoptotic signals of granulocyte-macrophage colony-stimulating factor are transduced via Jak2 tyrosine kinase in eosinophils. Eur J Immunol 1997;27:3536-9. [Crossref] [PubMed]
- Adachi T, Alam R. The mechanism of IL-5 signal transduction. Am J Physiol 1998;275:C623-33. [Crossref] [PubMed]
- Pazdrak K, Olszewska-Pazdrak B, Stafford S, et al. Lyn, Jak2, and Raf-1 kinases are critical for the antiapoptotic effect of interleukin 5, whereas only Raf-1 kinase is essential for eosinophil activation and degranulation. J Exp Med 1998;188:421-9. [Crossref] [PubMed]
- Pazdrak K, Stafford S, Alam R. The activation of the Jak-STAT 1 signaling pathway by IL-5 in eosinophils. J Immunol 1995;155:397-402. [Crossref] [PubMed]
- Quelle FW, Sato N, Witthuhn BA, et al. JAK2 associates with the beta c chain of the receptor for granulocyte-macrophage colony-stimulating factor, and its activation requires the membrane-proximal region. Mol Cell Biol 1994;14:4335-41. [Crossref] [PubMed]
- Tabèze L, Marchand-Adam S, Borie R, et al. Severe asthma with blood hypereosinophilia associated with JAK2 V617F mutation: a case series. Eur Respir J 2019;53:1802248. [Crossref] [PubMed]
- Zhang Y, Jing Y, Qiao J, et al. Activation of the mTOR signaling pathway is required for asthma onset. Sci Rep 2017;7:4532. [Crossref] [PubMed]
- Ma B, Athari SS, Mehrabi Nasab E, et al. PI3K/AKT/mTOR and TLR4/MyD88/NF-κB Signaling Inhibitors Attenuate Pathological Mechanisms of Allergic Asthma. Inflammation 2021;44:1895-907. [Crossref] [PubMed]
- Smith AK, Lange LA, Ampleford EJ, et al. Association of polymorphisms in CASP10 and CASP8 with FEV(1)/FVC and bronchial hyperresponsiveness in ethnically diverse asthmatics. Clin Exp Allergy 2008;38:1738-44. [PubMed]
- Karunakaran D, Nguyen MA, Geoffrion M, et al. RIPK1 Expression Associates With Inflammation in Early Atherosclerosis in Humans and Can Be Therapeutically Silenced to Reduce NF-κB Activation and Atherogenesis in Mice. Circulation 2021;143:163-77. [Crossref] [PubMed]
- Koren E, Fuchs Y. Modes of Regulated Cell Death in Cancer. Cancer Discov 2021;11:245-65. [Crossref] [PubMed]
- Webb LV, Barbarulo A, Huysentruyt J, et al. Survival of Single Positive Thymocytes Depends upon Developmental Control of RIPK1 Kinase Signaling by the IKK Complex Independent of NF-κB. Immunity 2019;50:348-361.e4. [Crossref] [PubMed]
- Yu J, Zhong B, Xiao Q, et al. Induction of programmed necrosis: A novel anti-cancer strategy for natural compounds. Pharmacol Ther 2020;214:107593. [Crossref] [PubMed]
- Inoue H, Hattori T, Zhou X, et al. Dysfunctional ErbB2, an EGF receptor family member, hinders repair of airway epithelial cells from asthmatic patients. J Allergy Clin Immunol 2019;143:2075-2085.e10. [Crossref] [PubMed]
- Inoue H, Akimoto K, Homma T, et al. Airway Epithelial Dysfunction in Asthma: Relevant to Epidermal Growth Factor Receptors and Airway Epithelial Cells. J Clin Med 2020;9:3698. [Crossref] [PubMed]
- Polosa R, Puddicombe SM, Krishna MT, et al. Expression of c-erbB receptors and ligands in the bronchial epithelium of asthmatic subjects. J Allergy Clin Immunol 2002;109:75-81. [Crossref] [PubMed]
- Huang JQ, Wang F, Wang LT, et al. Circular RNA ERBB2 Contributes to Proliferation and Migration of Airway Smooth Muscle Cells via miR-98-5p/IGF1R Signaling in Asthma. J Asthma Allergy 2021;14:1197-207. [Crossref] [PubMed]