
Dysregulated expression of microRNA involved in resistance to osimertinib in EGFR mutant non-small cell lung cancer cells
Highlight box
Key findings
• Our study found that miR-708-5p, miR-708-3p, miR-10395-3p, miR-7704 miR-34a-5p, miR-19b-1-5p, and miR-219a-5p may play key roles in osimertinib resistance.
What is known and what is new?
• A growing body of evidence has confirmed that altered miRNA expression is principally involved in the underlying mechanism of primary or even acquired resistance to TKIs.
• However, studies on the linkage between the altered miRNA expression and osimertinib resistance are few, and the effect of the miRNAs in this regard is still unclear. Thus, the aim of our study was to analyze the differential miRNAs in the acquired resistance to osimertinib.
What is the implication, and what should change now?
• The dysregulated expression of microRNA is involved in resistance to osimertinib in EGFR-mutant non-small cell lung cancer cells, and targeting altered miRNAs constitutes a promising strategy for overcoming osimertinib resistance.
Introduction
Lung cancer is the foremost cause of cancer-related incidence and mortality worldwide (1-3), with non-small cell lung cancer (NSCLC) accounting for approximately 85% of total lung cancer cases (1). Despite the progress in lung cancer detection and therapy, the 5-year overall survival rate remains poor (4). Epidermal growth factor receptor (EGFR) is the most common activating mutation in NSCLC, accounting for approximately 40–55% of cases in the Asian population and being associated with poor prognosis (5,6). Thus far, the small molecule tyrosine kinase inhibitors (TKIs), including erlotinib, gefitinib, afatinib, and osimertinib, have received approval in the standard first-line treatment of patients with EGFR-mutant NSCLC due to their proven benefits in the objective response rate (ORR), progression-free survival (PFS), and tolerability over cytotoxic chemotherapy (7-9). Moreover, osimertinib has shown superior curative effect compared to the earlier generation inhibitors. The Phase III FLAURA Randomized trial evaluated osimertinib as a first-line treatment that produced longer PFS and overall survival (OS) compared to first-generation TKIs (10,11). Consequently, osimertinib is the preferred first-line treatment for EGFR-positive NSCLC.
Despite of the satisfactory efficacy of osimertinib, acquired resistance inevitably occurs over time (12,13). Various molecular mechanisms of acquired resistance to osimertinib have been reported, including altered EGFR signaling pathway and activation of bypass pathways, such as RAS/RAF/MEK/ERK (MAPK), phosphatidylinositol 3-kinase/mammalian target of the rapamycin (mTOR), and MET/epidermal growth factor receptor 2 (HER2) amplification; RET fusion; and histological/phenotypic transformation (14-18). However, a consensus concerning these mechanisms has not yet been reached. Thus, further analysis on osimertinib resistance may be critical for determining suitable treatment strategies and improving prognosis.
MicroRNAs (miRNAs), a family of small noncoding RNA molecules of approximately 20–22 nucleotides in length, are essential to the regulation of multiple biological processes, including gene regulation, apoptosis, and maintenance of cell differentiation (19,20). In particular, miRNAs play a key role in regulating gene expression, and miRNAs are commonly dysregulated in cancer (21,22). In terms of lung cancer, miRNAs can be used as diagnostic and prognostic markers, and have a dual role in indicating carcinogenesis and behavior (23-25). Accumulating literature points to the altered expression of miRNAs as a key contributor to the underlying mechanism of primary and even acquired resistance to TKIs (26,27). For example, it has been found that the upregulation of miR-21 mediates gefitinib resistance in NSCLC via the activation of anaplastic lymphoma kinase (ALK)and signal-regulated kinase (ERK) pathway (28). In addition, previous studies have shown that miRNAs can restore the sensitivity of drug-resistant cells, thereby improving the therapeutic effect of EFGR-TKIs (29). Indeed, miR-133b was found to be associated with longer PFS when erlotinib was used as second- or third-line therapy for NSCLC (30).
However, studies on the linkage between the altered miRNA expression and resistance to osimertinib, a third-generation EGFR-TKI, are few, while the effect miRNAs in this process remains unclear (25,29,31). We thus hypothesized that differential expression of multiple miRNAs involves in the process of osimertinib resistance as drivers (32). Thus, the aim of our study was to analyze the miRNAs differences in acquired resistance to osimertinib. We present the following article in accordance with the MDAR reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-401/rc).
Methods
Cell culture
Two lung adenocarcinoma cell lines (A549 and NCI-H1975) were purchased from Procell Biologics Co., Ltd. (Wuhan, China) and H1975 from Beina Chuanglian Biotechnology Co., Ltd. (Beijing, China) cultured in MEM (Dulbecco’s Modified Eagle’s Medium) medium and Human Resources Processes Manual Introduction (HRPMI)-1640 medium containing 10% fetal bovine serum (FBS) and 1% penicillin and streptomycin respectively. The A549 cell line belong to the EGFR wild-type lung adenocarcinoma cell line. H1975 is a L858R mutant with a T790M mutant cell line. All cells were routinely screened for the absence of mycoplasma, and at least 3 independent experiments were performed for each condition.
Drug preparation for in vitro studies
AZD9291 (Osimertinib) was purchased from Aladdin’s official website , was dissolved in dimethyl sulfoxide (DMSO) to prepare 0.4-M stock solutions, which were aliquoted and stored at −80 ℃ and diluted as necessary for in vitro experiments.
Construction of osimertinib-resistant cell lines A549/Osi and H1975/Osi
The osimertinib-resistant cell lines were established using a stepwise method (33). Specifically, A549 and H1975 cell lines were cultured with increasing concentrations of AZD9291 starting at 0.5 mol/L. This was followed by a stepwise dose escalation every 7 days. After 7 months, A549 and H1975 cells could grow normally in 45 µmol/L and 50 µmol/L of AZD9291, respectively. The surviving cells were osimertinib-resistant cell lines and continued to be cultured in AZD9291-free medium for 2 weeks for subsequent studies.
Functional test of drug-resistant cell lines
Cell proliferation assay
Cell proliferation was assessed using cell counting kit 8 (CCK-8) assay to examine the effect of osimertinib on the 2 NSCLC cell lines. Cell suspensions were seeded in 96-well plate at a density of 3×103, and various concentrations of osimertinib were added. After incubation at 37 ℃ for 72 h, CCK-8 (Invigentech, Irvine, CA, USA) reagent was added to each well and further incubated for 2 h at 37 ℃. The absorbance of each well was measured at 450 nm using a microplate reader. The half maximal inhibitory concentration (IC50) value was defined as the concentration of AZD9291 needed for 50% reduction of growth. The corrected absorbance of each sample compared with that of the untreated control was calculated. Each experiment was performed independently 3 times.
Cell migration assay
We evaluated cell migration using scratch tests. In detail, the cells to be tested in the logarithmic growth stage were collected and seeded into a 6-well plate at a concentration of 3×105 cells/well, and the cell was scraped with a 100-µL sterile micropipette tip, Then, the serum-free medium was added to continue the culture. Finally, migratory cells were observed with an inverted fluorescence microscope (10×) at 0, 24, and 48 h and analyzed by using by Image-Pro Plus 6.0 software (Media Cybernetics, Rockville, MD, USA).
Colony formation assay
Cells (4×102) were seeded in 6-well plates and cultured for 2 weeks at the above-described conditions. Then, these plates were washed with phosphate-buffered saline (PBS) twice, and colonies were fixed with methanol for 10 min and stained with 0.1% crystal violet. Finally, colony numbers were counted using a microscope.
Transwell migration and invasion assay
We evaluated cell migration and invasion using scratch tests. Approximately 5×104 cells in serum-free medium were seeded in the upper chamber. Moreover, medium supplemented with FBS was filled into the bottom wells of the chamber. After incubation for 24 h, the cells that had not migrated were removed from the upper surface using cotton swabs, while cells that had invaded into the lower surface were fixed with paraformaldehyde and stained with crystal violet. The invasion assay also required the ABW matrix glue to be stored at −80 ℃ and placed in a refrigerator at 4 ℃ a day in advance to make it liquid. Serum-free medium and liquid substrate gel were thoroughly mixed at a dilution ratio of 1:8. The 100 µL diluted matrix gel was absorbed and added to the upper chamber of the Transwell chamber. The numbers of invasive cell were counted by capturing different fields using a microscope.
Bioinformatics analysis
First, miRNA sequencing was performed on the third generation EGFR-TKI-sensitive cell lines and the corresponding constructed drug-resistant cell lines. Second, miRDeep2 software was used to predict and identify new miRNA mature (star miRNA and mature miRNA) and precursor sequences of the species by combining homologous miRNA sequences of closely related species, including RNA fold and other RNA secondary structures, and the expression of new miRNA in each sample was counted. Thirdly. Deseq2 software was used to analyze the differentially expressed genes (DEGs) between the experimental group (drug-resistant cell line) and control group (normal cell lines), when meet statistical P value <0.05 and |log2 multiples (difference)| >1 conditions for the differences of miRNAs. Compared with normal cells, in drug-resistant strains, miRNAs that meet log2(difference multiple) >1 are up-regulated genes, and log2 (difference multiple) <−1 are down-regulated genes, while those that do not meet the above conditions are non-significant differentially expressed genes. Finally, miRNA target genes were predicted using miRanda and RNAhybrid, and the intersection of the 2 methods was taken as the final result. The R package “clusterProfiler” was used for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis based on all target genes of the differential miRNAs labeled as up or down in the differential expression analysis results. According to the PolymiRTS database, the Single Nucleotide Polymorphism Arrays (SNP) of the different known miRNAs were annotated. The above-described bioinformatic analysis steps were entrusted to Shanghai Tianhao Biotechnology Co., Ltd. (Shanghai, China) for completion.
Statistical analysis
All results are presented as the mean ± standard error of the mean (SEM) and were analyzed using SPSS 20.0 software (IBM Corp., Armonk, NY, USA). The standard Student t-test was used to determine the significance differences between the drug-resistant cell line and parent cell lines. P<0.05 denoted statistically significant difference.
Results
Morphological observations of drug-resistant cells
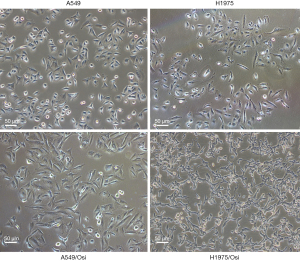
A morphological observation of drug-resistant cells is shown in Figure 1. Compared with the parent cells, drug-resistant cells tended to be spindle-like and irregularly shaped, with antennae, loss of epithelioid cell morphology, round vacuoles in cytoplasm, and an increase in the intercellular spaces with relatively slow growth (Figure 1).
Cell viability and proliferation analyses of resistant A549/Osi and H1975/Osi cells
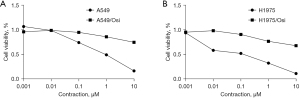
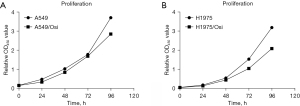
The cell viability and proliferation of resistant cells were measured with CCK8 assay. The results showed that the cell vitality of A549/Osi- and H1975/Osi-resistant cells was significantly increased (Figure 2), while the proliferation ability was slightly decreased and proliferation capacity compared with the parent cells (Figure 3). The mean IC50 values of A549, A549/Osi, H1975 and H1975/Osi cells were 0.093, 2.165, 0.022 and 2.445, respectively. Therefore, the resistance indexes (RI) of A549/Osi cells and H1975/Osi cells were 23.226 and 109.215, respectively. This promoted the successful construction of AZD9291 resistant cell lines (if RI >5, it is considered that drug-resistant cell lines meet the requirements of drug-resistant strains).
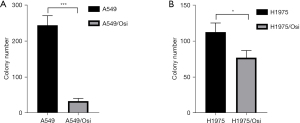
Clone formation analyses of resistant A549/Osi and H1975/Osi cells
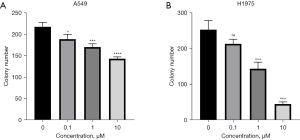
The results of the clone formation experiment showed that the colony number in the parent A549 and H1975 cells decreased with the increase in the concentration and thus in a dose-dependent manner (P<0.05) (Figure 4). In the absence of AZD9291, the number of colonies of A549/Osi and H1975/Osi cells was significantly lower than that of the parent cells (P<0.05) (Figure 5).
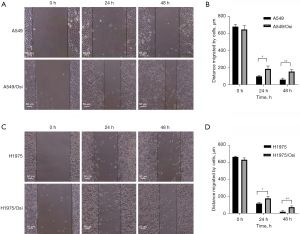
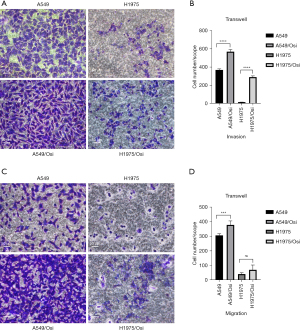
Migration and invasion capability analyses of resistant A549/Osi and H1975/Osi cells
Experiments on scratch (Figure 6) and Transwell experiments (Figure 7) were conducted to determine the migration and invasion capabilities of cells. The results revealed that the cell migration capacity of A549/Osi- and H1975/Osi-resistant cell was markedly increase compared with that in the parent cells in 24 and 48 h (P<0.05).
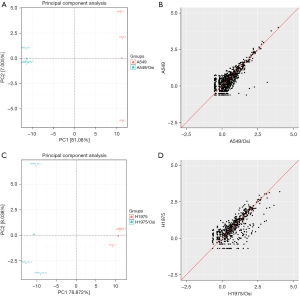
Differences in known miRNA expression in resistant A549/Osi and H1975/Osi cells
miRDeep2 software was used to compare the miRNA mature bodies in each sample and 2,656 miRNA expression levels were determined. Meanwhile, new miRNAs could be predicted and the expression levels of 1,420 potential new miRNAs were determined based on miRNA hairpin structure. By principal component analysis, it was found that samples in four different groups before and after drug resistance were well clustered, and all were grouped into one group respectively. The difference before and after drug resistance of A549 cell strain was greater than that before and after drug resistance of H1975 cell strain (Figure 8).
A total of 140 miRNAs were significantly differentially expressed in A549 cells, of which 64 were up-regulated and 76 were down-regulated after drug resistance (Figure 9). miR-708-5p, miR-516a-5p, miR-204-5p, miR-3085-3p and miR-708-3p ranked the top five in terms of up-regulated miRNA difference multiples. miR-3152-5p, miR-5000-5p, miR-122-3p, miR-122-5p, and miR-548w were the top five differences in the down-regulated miRNAs, respectively. In H1975 cells, 42 known miRNAs were differentially expressed before and after drug resistance. Compared with normal strains, 27 miRNAs were up-regulated and 15 were down-regulated in drug-resistant cells (Figure 9). Among them, miR-1268a, miR-7704, miR-4508, miR-10400-5p and miR-708-5p ranked top five in the order of up-regulated miRNA difference multiples. miR-127-3p, miR-19b-1-5p, miR-1-3p, miR-194-3p, and miR-25-5p ranked the top five differences in the down-regulated miRNA, respectively.
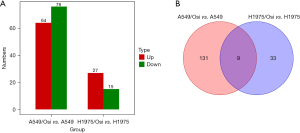
By combining the known miRNAs differentially expressed before and after drug resistance in the two cell lines, it was found that there were 9 miRNAs with significant differences in the two cell lines (Figure 9), and the differentially expressed trend of 7 miRNAs was consistent in the two cell lines. miR-708-5p, miR-708-3p, miR-10395-3p, miR-7704 and miR-34a-5p were significantly up-regulated in drug-resistant strains, while miR-19b-1-5p and miR-219a-5p were significantly down-regulated in drug-resistant strains. However, the differences between miR-1-3p and miR-138-5p were inconsistent.
Differential Gene Ontology (GO) enrichment in resistant A549/Osi and H1975/Osi cells
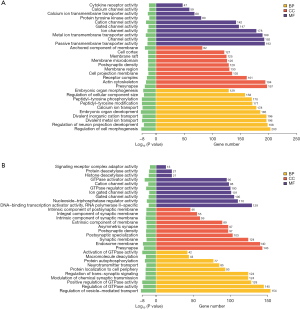
The target genes of differentially expressed miRNA were studied by GO and KEGG enrichment. By GO enrichment analysis, significantly differentially expressed genes were enriched according to biological process (BP), cell component (CC) and molecular function (MF). The most significantly enriched items are selected from the three categories of BP, CC, and MF respectively and shown in the figure (Figure 10).
Differential gene KEGG enrichment distribution plot
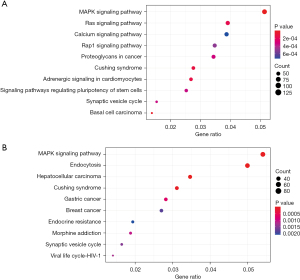
Scatter plots were used to show the results of KEGG enrichment, which were mainly measured by GeneRatio, P value and the number of genes enriched in this pathway. The results showed that MAPK signaling pathway was significant in both groups of cells (Figure 11).
SNP annotation results of known miRNAs
SNPs of miRNAs may affect miRNA-messenger RNA (mRNA) interactions, thereby altering gene expression and disease phenotype. In this study, 2 methods, miRanda and RNAhybrid, were used to predict miRNA target genes, with the intersection of the 2 methods being taken as the final result (Table 1).
Table 1
| miRNA | rs_id | Alleles | miRNA_position |
|---|---|---|---|
| hsa-miR-429 | rs370935426 | C/T | 4 |
| hsa-miR-429 | rs375039921 | C/T | 14 |
| hsa-miR-429 | rs368678282 | A/G | 21 |
| hsa-miR-548al | rs515924 | A/G | 8 |
| hsa-miR-548al | rs145394973 | A/T | 13 |
| hsa-miR-1304-3p | rs2155248 | A/C | −8 |
| hsa-miR-615-5p | rs375090644 | A/T | 13 |
| hsa-miR-625-3p | rs368340850 | C/G | 3 |
| hsa-miR-625-3p | rs12894182 | A/C | 19 |
| hsa-miR-3677-3p | rs138871259 | A/G | 4 |
| hsa-miR-3677-3p | rs373356017 | C/T | 3 |
| hsa-miR-122-3p | rs41292412 | C/T | 3 |
| hsa-miR-4746-5p | rs375980769 | G/T | 19 |
| hsa-miR-4746-5p | rs369043795 | C/T | 20 |
| hsa-miR-181d-5p | rs201654897 | A/G | 17 |
| hsa-miR-643 | rs371710100 | A/G | 1 |
| hsa-miR-516a-5p | rs368499496 | C/T | 20 |
| hsa-miR-1-3p | rs377171105 | G/T | 22 |
| hsa-miR-941 | rs111479365 | A/G | 12 |
| hsa-miR-941 | rs113283070 | C/G | 7 |
| hsa-miR-941 | rs113698386 | C/G | 15 |
| hsa-miR-941 | rs113309089 | A/G | 12 |
| hsa-miR-941 | rs113946605 | C/G | 15 |
| hsa-miR-941 | rs112466552 | A/G | 12 |
| hsa-miR-941 | rs34604519 | C/G | 15 |
| hsa-miR-941 | rs111993072 | A/G | 12 |
| hsa-miR-941 | rs113672516 | A/G | 6 |
| hsa-miR-941 | rs12625454 | C/G | 15 |
| hsa-miR-941 | rs111586745 | A/G | 12 |
| hsa-miR-941 | rs35544770 | A/G | 6 |
| hsa-miR-944 | rs190786647 | C/G | 16 |
| hsa-miR-944 | rs77473380 | C/T | 15 |
| hsa-miR-145-3p | rs190323149 | C/T | 6 |
| hsa-miR-7705 | rs3202310 | C/T | −1 |
| hsa-miR-188-5p | rs186369276 | G/T | 19 |
| hsa-miR-500a-5p | rs369176381 | G/T | 8 |
| hsa-miR-676-3p | rs376251880 | A/G | 17 |
| hsa-miR-664b-3p | rs112159031 | A/G | 8 |
miRNA, microRNA; SNP, single-nucleotide polymorphism.
Discussion
Osimertinib is currently the first choice for postoperative adjuvant therapy and advanced first-line treatment of EGFR-sensitive mutated NSCLC, and it is also the standard treatment for T790M mutation after first- and second-generation EGFR-TKI resistance. However, unfortunately, in most patients, resistance ultimately develops after an initial response to osimertinib, and the underlying mechanism of this resistance is complex. Most patients’ drug resistance mechanism is not clear, and there is no follow-up precise treatment, with the prognosis being poor (34,35).
Individual miRNAs play a key role in targeting and regulating the expression of numerous mRNAs either by promoting mRNA degradation or inhibiting their translation (21). Aberrant miRNAs expression patterns can be used as diagnostic and prognostic markers in NSCLC (36). A large number of studies have reported the role of miRNAs in cancer progression, such as proliferation, invasion, metastasis, metabolism, and drug resistance (24,37-40). Importantly, studies have identified that cancer cells develop drug resistance via multiple complex mechanisms that involve in various miRNAs by intervening in cell-cell interaction (26,27). In this study, through the biosynthesis analysis, we identified the differential expression patterns among miRNAs after cell resistance. Specifically, the expression of miR-708-5p, miR-708-3p, miR-10395-3p, miR-7704, and miR-34a-5p was upregulated while the expression of miR-19b-1-5p and miR-219a-5p was downregulated.
Other research has demonstrated that miR-708 suppresses proliferation, survival, and migration of lung cancer cells (41). Specifically, chemoresistant cells were found to reduce miR-708-5p expression compared to naïve lung cancer cells, and combination treatment with miR-708-5p was shown to partially restore the antiproliferative effects of chemotherapy and fully restored its proapoptotic qualities. Findings from our study were inconsistent with these results, and may possibly be explained by the following. First, different treatments were involved in the different proresistance pathways against chemoresistant and TKI-resistant cells. Second, the same miRNAs may have different effects that are modulated in a specific setting depending on their direct targets and the activated downstream signaling pathways. Finally, there also may other stress-associated transcription factors promoting miR-708-5p expression.
To date, several lines of evidence have shown that miR-34 and miR-219 regulate the survival and motility of cancer cells by activating different multiple targets. miR-34 and miR-219 downregulation is a frequently observed in NSCLC cell lines (42,43). However, our study is the first of its kind to investigate the potential association between miR-219, miR-34, and osimertinib resistance. Therefore, although current studies lack relevant functional assays, it is reasonable to speculate that resistance effects of miRNAs can be combined.
In this study, 7 miRNAs were found to be associated with osimertinib resistance in A549 and H1975 cell lines according to biosynthesis analysis. miR-708-5p, has been shown to repress pro-resistant signaling pathways, including COX-2 and mPGES-1. Moreover, combination treatment of erlotinib (ERL) or paclitaxel (PAC) with miR-708-5p enhances COX-2 and mPGES-1 protein suppression. combination chemotherapeutic and miR-708-5p treatment intensifies the anti-proliferative and pro-apoptotic effects of ERL and PAC (41). It has been confirmed that the regulation of EMT inhibition mediated by mIR-34a-5P-and HOTAIR may provide a new therapeutic paradigm for lung cancer (44). The reviews of the relevant literature points to miR-708-5p and miR-34a-5p as having particular importance (45). Therefore, miR-708-5p and miR-34a-5p need to be further analyzed. Further research directions relevant to this study include comparing the expression of serum miRNAs in patients with EGFR-sensitive mutant NSCLC before and after osimertinib resistance, and the possibility of animal experiments to determine the role of miRNAs in drug resistance after intervention; whether the tumor can regain its sensitivity to osimertinib.
Conclusions
In A549 and H1975 cell lines, osimertinib resistance was associated with the upregulation of miR-708-5p, miR-708-3p, miR-10395-3p, miR-7704, and miR-34a-5p expression, and the downregulation of miR-19b-1-5p and miR-219a-5p expression. These results, in combination with the relevant literature, suggest that miR-708-5p and miR-34a-5p may play a tumor-promoting role in NSCLC and be positively correlated with osimertinib resistance.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-401/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-401/dss
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-401/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-401/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Schabath MB, Cote ML. Cancer Progress and Priorities: Lung Cancer. Cancer Epidemiol Biomarkers Prev 2019;28:1563-79. [Crossref] [PubMed]
- The Lancet. Lung cancer: some progress, but still a lot more to do. Lancet 2019;394:1880. [Crossref]
- Siegel RL, Miller KD, Wagle NS, et al. Cancer statistics, 2023. CA Cancer J Clin 2023;73:17-48. [Crossref] [PubMed]
- Duma N, Santana-Davila R, Molina JR. Non-Small Cell Lung Cancer: Epidemiology, Screening, Diagnosis, and Treatment. Mayo Clin Proc 2019;94:1623-40. [Crossref] [PubMed]
- Midha A, Dearden S, McCormack R. EGFR mutation incidence in non-small-cell lung cancer of adenocarcinoma histology: a systematic review and global map by ethnicity (mutMapII). Am J Cancer Res 2015;5:2892-911. [PubMed]
- Shi Y, Au JS, Thongprasert S, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER). J Thorac Oncol 2014;9:154-62. [Crossref] [PubMed]
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. [Crossref] [PubMed]
- Yang JJ, Zhou Q, Yan HH, et al. A phase III randomised controlled trial of erlotinib vs gefitinib in advanced non-small cell lung cancer with EGFR mutations. Br J Cancer 2017;116:568-74. [Crossref] [PubMed]
- Sequist LV, Yang JC, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol 2013;31:3327-34. [Crossref] [PubMed]
- Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:113-25. [Crossref] [PubMed]
- Ramalingam SS, Vansteenkiste J, Planchard D, et al. Overall Survival with Osimertinib in Untreated, EGFR-Mutated Advanced NSCLC. N Engl J Med 2020;382:41-50. [Crossref] [PubMed]
- Thai AA, Solomon BJ, Sequist LV, et al. Lung cancer. Lancet 2021;398:535-54. [Crossref] [PubMed]
- Gomatou G, Syrigos N, Kotteas E. Osimertinib Resistance: Molecular Mechanisms and Emerging Treatment Options. Cancers (Basel) 2023;15:841. [Crossref] [PubMed]
- Leonetti A, Sharma S, Minari R, et al. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br J Cancer 2019;121:725-37. [Crossref] [PubMed]
- He J, Huang Z, Han L, et al. Mechanisms and management of 3rd-generation EGFR-TKI resistance in advanced non-small cell lung cancer Int J Oncol 2021;59:90. (Review). [Crossref] [PubMed]
- Huang WC, Yadav VK, Cheng WH, et al. The MEK/ERK/miR-21 Signaling Is Critical in Osimertinib Resistance in EGFR-Mutant Non-Small Cell Lung Cancer Cells. Cancers (Basel) 2021;13:6005. [Crossref] [PubMed]
- Yu J, Zhang L, Peng J, et al. Dictamnine, a novel c-Met inhibitor, suppresses the proliferation of lung cancer cells by downregulating the PI3K/AKT/mTOR and MAPK signaling pathways. Biochem Pharmacol 2022;195:114864. [Crossref] [PubMed]
- Shah MP, Neal JW. Targeting Acquired and Intrinsic Resistance Mechanisms in Epidermal Growth Factor Receptor Mutant Non-Small-Cell Lung Cancer. Drugs 2022;82:649-62. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [Crossref] [PubMed]
- Vishnoi A, Rani S. miRNA Biogenesis and Regulation of Diseases: An Updated Overview. Methods Mol Biol 2023;2595:1-12. [Crossref] [PubMed]
- Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med 2014;20:460-9. [Crossref] [PubMed]
- Hua X, Li Y, Pentaparthi SR, et al. Landscape of MicroRNA Regulatory Network Architecture and Functional Rerouting in Cancer. Cancer Res 2023;83:59-73. [Crossref] [PubMed]
- Wu KL, Tsai YM, Lien CT, et al. The Roles of MicroRNA in Lung Cancer. Int J Mol Sci 2019;20:1611. [Crossref] [PubMed]
- Iqbal MA, Arora S, Prakasam G, et al. MicroRNA in lung cancer: role, mechanisms, pathways and therapeutic relevance. Mol Aspects Med 2019;70:3-20. [Crossref] [PubMed]
- Wang H, Wang L, Sun G. MiRNA and Potential Prognostic Value in Non-Smoking Females with Lung Adenocarcinoma by High-Throughput Sequencing. Int J Gen Med 2023;16:683-96. [Crossref] [PubMed]
- Hassanein SS, Ibrahim SA, Abdel-Mawgood AL. Cell Behavior of Non-Small Cell Lung Cancer Is at EGFR and MicroRNAs Hands. Int J Mol Sci 2021;22:12496. [Crossref] [PubMed]
- Ma X, Liang AL, Liu YJ. Research progress on the relationship between lung cancer drug-resistance and microRNAs. J Cancer 2019;10:6865-75. [Crossref] [PubMed]
- Jing C, Cao H, Qin X, et al. Exosome-mediated gefitinib resistance in lung cancer HCC827 cells via delivery of miR-21. Oncol Lett 2018;15:9811-7. [Crossref] [PubMed]
- He B, Zhao Z, Cai Q, et al. miRNA-based biomarkers, therapies, and resistance in Cancer. Int J Biol Sci 2020;16:2628-47. [Crossref] [PubMed]
- Bisagni A, Pagano M, Maramotti S, et al. Higher expression of miR-133b is associated with better efficacy of erlotinib as the second or third line in non-small cell lung cancer patients. PLoS One 2018;13:e0196350. [Crossref] [PubMed]
- Li X, Chen C, Wang Z, et al. Elevated exosome-derived miRNAs predict osimertinib resistance in non-small cell lung cancer. Cancer Cell Int 2021;21:428. [Crossref] [PubMed]
- Janpipatkul K, Trachu N, Watcharenwong P, et al. Exosomal microRNAs as potential biomarkers for osimertinib resistance of non-small cell lung cancer patients. Cancer Biomark 2021;31:281-94. [Crossref] [PubMed]
- Tanaka K, Yu HA, Yang S, et al. Targeting Aurora B kinase prevents and overcomes resistance to EGFR inhibitors in lung cancer by enhancing BIM- and PUMA-mediated apoptosis. Cancer Cell 2021;39:1245-1261.e6. [Crossref] [PubMed]
- Schmid S, Li JJN, Leighl NB. Mechanisms of osimertinib resistance and emerging treatment options. Lung Cancer 2020;147:123-9. [Crossref] [PubMed]
- Zeng Y, Yu D, Tian W, et al. Resistance mechanisms to osimertinib and emerging therapeutic strategies in nonsmall cell lung cancer. Curr Opin Oncol 2022;34:54-65. [Crossref] [PubMed]
- Lamb YN. Osimertinib: A Review in Previously Untreated, EGFR Mutation-Positive, Advanced NSCLC. Target Oncol 2021;16:687-95. [Crossref] [PubMed]
- Li B, Cao Y, Sun M, et al. Expression, regulation, and function of exosome-derived miRNAs in cancer progression and therapy. FASEB J 2021;35:e21916. [Crossref] [PubMed]
- Zhong S, Golpon H, Zardo P, et al. miRNAs in lung cancer. A systematic review identifies predictive and prognostic miRNA candidates for precision medicine in lung cancer. Transl Res 2021;230:164-96. [Crossref] [PubMed]
- Catellani C, Ravegnini G, Sartori C, et al. GH and IGF System: The Regulatory Role of miRNAs and lncRNAs in Cancer. Front Endocrinol (Lausanne) 2021;12:701246. [Crossref] [PubMed]
- Dragomir MP, Knutsen E, Calin GA. Classical and noncanonical functions of miRNAs in cancers. Trends Genet 2022;38:379-94. [Crossref] [PubMed]
- Monteleone NJ, Lutz CS. miR-708-5p enhances erlotinib/paclitaxel efficacy and overcomes chemoresistance in lung cancer cells. Oncotarget 2020;11:4699-721. [Crossref] [PubMed]
- Kim YH, Lee WK, Lee EB, et al. Combined Effect of Metastasis-Related MicroRNA, miR-34 and miR-124 Family, Methylation on Prognosis of Non-Small-Cell Lung Cancer. Clin Lung Cancer 2017;18:e13-20. [Crossref] [PubMed]
- Daugaard I, Knudsen A, Kjeldsen TE, et al. The association between miR-34 dysregulation and distant metastases formation in lung adenocarcinoma. Exp Mol Pathol 2017;102:484-91. [Crossref] [PubMed]
- Zheng F, Li J, Ma C, et al. Novel regulation of miR-34a-5p and HOTAIR by the combination of berberine and gefitinib leading to inhibition of EMT in human lung cancer. J Cell Mol Med 2020;24:5578-92. [Crossref] [PubMed]
- Chai R, Xu C, Lu L, et al. Quercetin inhibits proliferation of and induces apoptosis in non-small-cell lung carcinoma via the lncRNA SNHG7/miR-34a-5p pathway. Immunopharmacol Immunotoxicol 2021;43:693-703. [Crossref] [PubMed]
(English Language Editor: J. Gray)


