
Overexpressed FOXM1 collaborates with MMB to increase WEE1 inhibitor sensitivity in NSCLC
Highlight box
Key findings
• The FOXM1-MMB complex was found to be necessary for WEE1 inhibitor sensitivity.
• FOXM1 upregulation enhanced DNA replication stress.
• The FOXM1-MMB complex activated G2/M checkpoints.
What is known and what is new?
• According to one study, MMB-FOXM1-driven premature mitosis is necessary for sensitivity to CHK1 inhibitors.
• This is the first study to discover that overexpressed FOXM1 collaborates with MMB to increase WEE1 inhibitor sensitivity in NSCLC.
What is the implication, and what should change now?
• This work revealed that the complex formed by overexpressed FOXM1 and MMB might be used as a potential therapeutic biomarker for NSCLC.
Introduction
More than 80% of all lung cancers are non-small cell lung cancer (NSCLC), and this disease is a major cause of cancer deaths worldwide (1-3). Although great improvements have been made due to progress in treatment strategies and surgical techniques, the 5-year survival rate is only slightly higher than 15% (4,5). And, the prognosis of NSCLC patients is discouraging. Currently, effective treatment strategies such as radiotherapy and chemotherapy are limited. More and more molecular biomarkers have been verified to participate into the progression of NSCLC. Therefore, identifying novel specific molecular biomarkers is of great importance to improve the treatment of NSCLC.
The transcription factor forkhead box M1 (FOXM1) is a well-known oncogene and an essential factor for cell proliferation in cancers (6). For example, FOXM1 facilitates breast cancer cell stemness and migration in a Yes-associated protein 1 (YAP1)-dependent manner (7). High expression of FOXM1 is critical for sustaining cell proliferation in mitochondrial DNA-less liver cancer cells (8). FOXM1 promotes the migration of ovarian cancer cells through Keratin 5 (KRT5) and Keratin 5 (KRT7) (9). A novel FOXM1-proteasome subunit beta type-4 (PSMB4) axis contributes to the proliferation and progression of cervical cancer (10).
Regulation of the cell cycle is critical for normal development; cell cycle disruption causes a number of diseases, especially cancers (11,12). FOXM1 is a pivotal modulator of periodic gene transcription in the G2-M phase of the cell cycle, and thus, it seems to be connected with the increased cell proliferation ability of tumors (13-15). MuvB is an evolutionarily conserved multisubunit complex, and it can modulate gene expression during the cell cycle (16,17). During S phase, the interaction between the MuvB core and p130/E2F4/DP1 is abolished, and MuvB then interacts with the B-MYB (V-Myb avian myeloblastosis viral oncogene homolog-like 2, MYBL2) transcription factor to form the MYB-MuvB complex (also called MMB) (18,19).
The complex formed by MMB and FOXM1 (MMB-FOXM1) plays a vital role in cell cycle progression by modulating the transcription of genes needed for cytokinesis and mitosis (20). In the cell cycle, the E2F-dependent wave of gene expression at the G1/S transition accelerates DNA replication, and the MMB-FOXM1 complex drives the second wave of gene expression at the G2/M transition, subsequently boosting mitosis (21,22). FOXM1 is then recruited to the promoters of these genes through an MMB-dependent pattern (19). This recruitment is consistent with the increased levels of hundreds of G2/M cell cycle genes, such as Cell Division Cycle Protein 25B (CDC25B), Cyclin A2 (CCNA2), Polo-like Kinase 1 (PLK1) and Cyclin B1 (CCNB1) (23,24). According to one study, MMB-FOXM1-driven premature mitosis is necessary for sensitivity to checkpoint kinase 1 (CHK1) inhibitors (25). However, the regulatory role of the MMB-FOXM1 complex in NSCLC keeps unclear, and needs more exploration in NSCLC.
Wee1-like protein kinase (WEE1) negatively modulates CDK1, and inhibition of WEE1 eliminates inhibitory phosphorylation of CDK1, thereby inducing tumor cell apoptosis (26). This regulation of WEE1 inhibitor also exist in NSCLC, that Wee1 inhibitor MK1775 strengthens sorafenib sensibility in KRAS mutated NSCLC cells (27). In addition, WEE1 inhibitor AZD1775 has improvement effects on LKB1-deficient NSCLC (28). Besides, WEE1 inhibitor enhances chemosensitivity in EGFR-TKIs resistant NSCLC (29). However, the relationship between MMB-FOXM1 complex and WEE1 inhibitor in NSCLC remain vague.
This study aimed to explore the relationship between FOXM1 overexpression and WEE1 inhibitor sensitivity in NSCLC. Our findings revealed that overexpressed FOXM1 collaborates with MMB to increase WEE1 inhibitor sensitivity in NSCLC. This discovery might provide novel insight into identifying new biomarkers for NSCLC treatment. We present this article in accordance with the MDAR reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-750/rc).
Methods
Cell lines and cell culture
The NSCLC cell lines H1650 (TCHu152) and H2228 (SCSP-5001) were acquired from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cells were maintained in Dulbecco’s modified Eagle’s medium (C11995500BT, DMEM, Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (SH30068.03, FBS, HyClone, Logan, UT, USA), penicillin (100 units/mL) and streptomycin (100 mg/mL) at 37 ℃ with 5% CO2.
Transfection
The FOXM1 overexpression vector (pcDNA3.1-FOXM1) and small interfering RNAs (siRNAs) targeting FOXM1 and MMB (siFOXM1-1, siFOXM1-2, siFOXM1-3, siLIN54-1, siLIN54-2, siLIN54-3) were obtained from GenePharma (Shanghai, China). These vectors were transfected into NSCLC cells with Lipofectamine 2000 (Invitrogen, USA). AZD-1775 (the WEE1 inhibitor, 0.5 µM, SC6677) was purchased from Beyotime (Shanghai, China).
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
The RT-qPCR was performed to examining mRNA expressions. TRIzol reagent (15596018, Thermo Fisher Scientific) was employed to isolate RNA from NSCLC cells. Reverse transcription reagents (Applied Biosystems, Foster City, USA) were applied for cDNA synthesis by reverse transcription. RT-qPCR was conducted with SYBR Green (Takara Bio, Japan), and GAPDH was utilized as an internal control. The relative expression levels were calculated through the 2−ΔΔCt method.
Western blot
The western blot was performed to measuring protein expressions. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was applied for isolation of proteins, which were then transferred to polyvinylidene fluoride (IPVH00010, PVDF, Millipore, Bedford, MA, USA) membranes. After blocking with nonfat milk, the membranes were incubated with primary antibodies at 4 ℃ overnight. After washing, the membrane was incubated with goat anti-rabbit IgG (ab6721, 1: 2,000, Abcam) as the secondary antibody, and the bands were visualized with an enhanced chemiluminescence detection system. The primary antibodies included anti-FOXM1 (ab180710, Abcam), anti-LIN54 (ab138425, Abcam), anti-RPA (ab2175, Abcam), anti-γH2AX (ab11175, Abcam), anti-CCNB (ab32053, Abcam) and anti-β-Actin (ab6276, Abcam).
Cell Counting Kit-8 (CCK-8) assay
The CCK-8 assay was performed to measuring cell survival. NSCLC cells were plated into 96-well plates. The cells were incubated for 24 h and then supplemented with AZD-1775 for 24 h. Next, 10 mL of CCK-8 reagent was mixed into each well. After 2 h, the optical density (OD =450 nm) was assessed with an enzyme immunoassay analyzer (Thermo Fisher Scientific).
Statistical analysis
GraphPad Prism software, version 8.0 (GraphPad Software, La Jolla, CA) was utilized for statistical analysis. All experiments were performed in triplicate with randomization, and data are expressed as the mean ± standard deviation (SD) values. Two-group (or multiple-group) comparisons were carried out with Student’s t test [or one-way variance analysis (ANOVA)]. All statistical tests were two-sided, and statistical significance was set at P<0.05.
Results
The FOXM1-MMB complex was found to be necessary for WEE1 inhibitor sensitivity
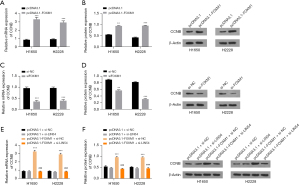
First, FOXM1 was overexpressed or silenced using the appropriate vectors to verify the role of FOXM1 in WEE1 inhibitor sensitivity. The mRNA and protein expression levels of FOXM1 were increased by the FOXM1 overexpression vector (Figure 1A,1B). Additionally, the mRNA and protein expression levels of FOXM1 were decreased by the FOXM1 siRNAs (Figure 1C,1D). After treatment with AZD-1775 (WEE1 inhibitor), cell survival was obviously decreased by either overexpression or inhibition of FOXM1 (Figure 1E,1F). The MMB complex is formed by MYBL2 and MuvB, while MuvB is composed of LIN54, LIN9, LIN37, LIN52 and RBBP4. LIN54 plays a crucial role in the formation of the MMB complex, and the influence of MMB was eliminated by silencing LIN54 expression. The mRNA and protein expression of LIN54 was decreased by silencing LIN54 (Figure 1G-1H). The protein expression of FOXM1 and LIN54 were not changed after AZD-1775 treatment (Figure 1I). Furthermore, after AZD-1775 treatment, the decrease in cell survival mediated by FOXM1 overexpression could be reversed by LIN54 knockdown (Figure 1J). In addition, the cell survival in the control group did not differ obviously from that in the pcDNA3.1-FOXM1+siLIN54 group, indicating that overexpression of FOXM1 increased WEE1 inhibitor sensitivity in a manner dependent on the complex formed with MMB. Taken together, these findings indicate that the FOXM1-MMB complex is necessary for WEE1 inhibitor sensitivity.

FOXM1 upregulation enhanced DNA replication stress
Next, experiments were performed to confirm the role of FOXM1 in DNA replication stress and DNA damage. Replication protein A (RPA) is a marker of replicative stress, and γH2AX is a marker of DNA double-strand breaks. As displayed in Figure 2A,2B, the mRNA and protein levels of RPA were increased by overexpressing FOXM1, but that of γH2AX did not obviously change. Moreover, FOXM1 overexpression enhanced the mRNA and protein expression of RPA but did not affect the expression of γH2AX (Figure 2C,2D). We found that the mRNA and protein expression levels of RPA and γH2AX were increased after AZD-1775 treatment and FOXM1 overexpression (Figure 2E,2F). In addition, the mRNA and protein expression of RPA was downregulated and that of γH2AX was not changed after AZD-1775 treatment and FOXM1 knockdown (Figure 2G,2H). In summary, FOXM1 upregulation enhanced DNA replication stress.

The FOXM1-MMB complex activated G2/M checkpoints
CCNB and CDK1 form a complex to regulate the G2/M checkpoint. FOXM1 can enhance CCNB expression and increase the threshold content of the CCNB-CDK1 complex, facilitating the opening of the G2/M checkpoint. The CCNB mRNA and protein expression levels were increased after upregulating FOXM1 (Figure 3A,3B). Additionally, the CCNB mRNA and protein expression levels were decreased after silencing FOXM1 (Figure 3C,3D). This finding suggested that overexpression of FOXM1 enhances CCNB expression and increases the threshold content of the CCNB-CDK1 complex. Finally, it was demonstrated that the increased mRNA and protein expression of CCNB mediated by FOXM1 could be rescued by silencing LIN54 (Figure 3E,3F). Furthermore, CCNB expression in the pcDNA3.1+si-NC group showed no obvious difference from that in the pcDNA3.1-FOXM1+siLIN54 group, indicating that FOXM1 and MMB form a complex to regulate the CCNB expression level. These findings indicated that the FOXM1-MMB complex activated G2/M checkpoints.
Discussion
The regulatory mechanisms of cell cycle checkpoints and DNA repair have been the focus of many studies (30,31). However, clinical translation of the findings has been unsatisfactory. A critical challenge is the lack of validated predictive biotargets. These biotargets can easily identify tumors that may respond to DNA repair pathway inhibitors, particularly inhibitors of DNA replication stress (32,33).
The G2/M cell cycle checkpoint plays a key role in the cell cycle to guarantee that DNA repair is finished before entry into mitosis (34). Cyclin dependent kinase 1 (CDK1) is a crucial regulatory factor of the G2/M transition, and its phosphorylation controls whether cells enter mitosis (35). WEE1 serves as a negative modulator of CDK1, and WEE1 suppression eliminates inhibitory phosphorylation of CDK1 (36). When cells enter mitosis, the continuous accumulation of damaged DNA induces apoptosis, leading to tumor cell death (37,38).
One inhibitor, AZD1775, targets WEE1 kinase, which is an important regulator of the G2/M checkpoint (39,40). AZD1775 negatively modulates mitosis by inactivating CDK1, inducing G2/M arrest and permitting DNA repair (41,42). Thus, cells with DNA replication stress are susceptible to WEE1 kinase inhibition (43). In view of the complex modulation of cell cycle checkpoints, it is necessary to identify alternative pathways and biomarkers that can help predict the response to WEE1 kinase inhibition.
Cyclin E upregulation in triple-negative breast cancer enhances sensitivity to WEE1 inhibitors (44). Is there any association between FOXM1 expression and WEE1 inhibitor sensitivity in NSCLC? The MMB complex is formed by MYBL2 and MuvB, while MuvB is composed of LIN54, LIN9, LIN37, LIN52, and RBBP4 (16,45). LIN54 plays a crucial role in the formation of the MMB complex, and silencing of LIN54 can reduce the effect of MMB. Our work demonstrated that after AZD-1775 treatment, the decrease in cell survival mediated by FOXM1 overexpression could be reversed by LIN54 knockdown and that cell survival in the control group did not differ obviously from that in the pcDNA3.1-FOXM1+siLIN54 group, indicating that the FOXM1-MMB complex was necessary for WEE1 inhibitor sensitivity. Moreover, the mRNA and protein expression of replicative stress marker-RPA and DNA double-strand break marker-γH2AX were increased after AZD-1775 treatment and FOXM1 overexpression, suggesting that FOXM1 upregulation enhanced DNA replication stress and DNA damage. Finally, we found that the increased mRNA and protein expression of CCNB mediated by FOXM1 could be rescued by silencing LIN54 and that CCNB expression in the control group did not differ obviously from that in the pcDNA3.1-FOXM1+siLIN54 group. These findings revealed that the FOXM1-MMB complex activated G2/M checkpoints.
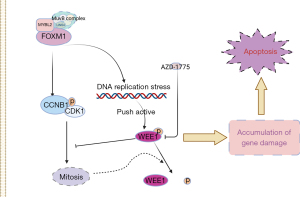
As shown in Figure 4, FOXM1 overexpression increased DNA replication stress, which increased DNA replication and pressure on the WEE1 checkpoint. On the other hand, FOXM1 can enhance CCNB expression, increase the threshold content of the CCNB/CDK1 complex, facilitate mitosis, and promote WEE1 dephosphorylation. Under these two effects, sensitivity to the WEE1 inhibitor AZD-1775 is increased, which leads to the accumulation of DNA damage and drives the activation of apoptosis.
Conclusions
In this study, we verified that overexpression of FOXM1 induces DNA replication stress and stimulates DNA repair responses, thereby sensitizing cells to AZD1775. Thus, this work revealed that the complex formed by overexpressed FOXM1 and MMB might be used as a potential therapeutic biomarker for NSCLC, making NSCLC treatment decisions more accurate.
Acknowledgments
Funding: This work was supported by the Wu Jieping Medical Foundation (No. 320.6750.15047, to Keqiang Liu).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-750/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-750/dss
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-750/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-750/coif). Keqiang Liu reports that this work was supported by the Wu Jieping Medical Foundation (No. 320.6750.15047). The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553:446-54. [Crossref] [PubMed]
- Chen P, Liu Y, Wen Y, et al. Non-small cell lung cancer in China. Cancer Commun (Lond) 2022;42:937-70. [Crossref] [PubMed]
- Arbour KC, Riely GJ. Systemic Therapy for Locally Advanced and Metastatic Non-Small Cell Lung Cancer: A Review. JAMA 2019;322:764-74. [Crossref] [PubMed]
- He J, Li Y, An J, et al. Surgical treatment in non-small cell lung cancer with pulmonary oligometastasis. World J Surg Oncol 2017;15:36. [Crossref] [PubMed]
- Gartel AL. FOXM1 in Cancer: Interactions and Vulnerabilities. Cancer Res 2017;77:3135-9. [Crossref] [PubMed]
- Sun HL, Men JR, Liu HY, et al. FOXM1 facilitates breast cancer cell stemness and migration in YAP1-dependent manner. Arch Biochem Biophys 2020;685:108349. [Crossref] [PubMed]
- Higurashi M, Maruyama T, Nogami Y, et al. High expression of FOXM1 critical for sustaining cell proliferation in mitochondrial DNA-less liver cancer cells. Exp Cell Res 2020;389:111889. [Crossref] [PubMed]
- Zhang Z, Tu K, Liu F, et al. FoxM1 promotes the migration of ovarian cancer cell through KRT5 and KRT7. Gene 2020;757:144947. [Crossref] [PubMed]
- Zhou DM, Liu J, Liu F, et al. A novel FoxM1-PSMB4 axis contributes to proliferation and progression of cervical cancer. Biochem Biophys Res Commun 2020;521:746-52. [Crossref] [PubMed]
- Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature 2004;432:316-23. [Crossref] [PubMed]
- Williams GH, Stoeber K. The cell cycle and cancer. J Pathol 2012;226:352-64. [Crossref] [PubMed]
- Wierstra I, Alves J. FOXM1, a typical proliferation-associated transcription factor. Biol Chem 2007;388:1257-74. [Crossref] [PubMed]
- Wierstra I. The transcription factor FOXM1 (Forkhead box M1): proliferation-specific expression, transcription factor function, target genes, mouse models, and normal biological roles. Adv Cancer Res 2013;118:97-398. [Crossref] [PubMed]
- Takei J, Tanaka T, Teshigawara A, et al. Alteration of FOXM1 expression and macrophage polarization in refractory meningiomas during long-term follow-up. Transl Cancer Res 2021;10:553-66. [Crossref] [PubMed]
- Iness AN, Litovchick L, Muv B. A Key to Cell Cycle Control in Ovarian Cancer. Front Oncol 2018;8:223. [Crossref] [PubMed]
- Mowla SN, Lam EW, Jat PS. Cellular senescence and aging: the role of B-MYB. Aging Cell 2014;13:773-9. [Crossref] [PubMed]
- Fischer M, Müller GA. Cell cycle transcription control: DREAM/MuvB and RB-E2F complexes. Crit Rev Biochem Mol Biol 2017;52:638-62. [Crossref] [PubMed]
- Sadasivam S, Duan S, DeCaprio JA. The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev 2012;26:474-89. [Crossref] [PubMed]
- Fischer M, Grossmann P, Padi M, et al. Integration of TP53, DREAM, MMB-FOXM1 and RB-E2F target gene analyses identifies cell cycle gene regulatory networks. Nucleic Acids Res 2016;44:6070-86. [Crossref] [PubMed]
- Wolter P, Hanselmann S, Pattschull G, et al. Central spindle proteins and mitotic kinesins are direct transcriptional targets of MuvB, B-MYB and FOXM1 in breast cancer cell lines and are potential targets for therapy. Oncotarget 2017;8:11160-72. [Crossref] [PubMed]
- Tanaka Y, Patestos NP, Maekawa T, et al. B-myb is required for inner cell mass formation at an early stage of development. J Biol Chem 1999;274:28067-70. [Crossref] [PubMed]
- Cheng XH, Black M, Ustiyan V, et al. SPDEF inhibits prostate carcinogenesis by disrupting a positive feedback loop in regulation of the Foxm1 oncogene. PLoS Genet 2014;10:e1004656. [Crossref] [PubMed]
- Wiseman EF, Chen X, Han N, et al. Deregulation of the FOXM1 target gene network and its coregulatory partners in oesophageal adenocarcinoma. Mol Cancer 2015;14:69. [Crossref] [PubMed]
- Branigan TB, Kozono D, Schade AE, et al. MMB-FOXM1-driven premature mitosis is required for CHK1 inhibitor sensitivity. Cell Rep 2021;34:108808. [Crossref] [PubMed]
- Matheson CJ, Backos DS, Reigan P. Targeting WEE1 Kinase in Cancer. Trends Pharmacol Sci 2016;37:872-81. [Crossref] [PubMed]
- Caiola E, Frapolli R, Tomanelli M, et al. Wee1 inhibitor MK1775 sensitizes KRAS mutated NSCLC cells to sorafenib. Sci Rep 2018;8:948. [Crossref] [PubMed]
- Richer AL, Cala JM, O'Brien K, et al. WEE1 Kinase Inhibitor AZD1775 Has Preclinical Efficacy in LKB1-Deficient Non-Small Cell Lung Cancer. Cancer Res 2017;77:4663-72. [Crossref] [PubMed]
- Liu D, Cao Z, Xu W, et al. Enhancement of chemosensitivity by WEE1 inhibition in EGFR-TKIs resistant non-small cell lung cancer. Biomed Pharmacother 2019;117:109185. [Crossref] [PubMed]
- Barnum KJ, O'Connell MJ. Cell cycle regulation by checkpoints. Methods Mol Biol 2014;1170:29-40. [Crossref] [PubMed]
- Yasui H, Iizuka D, Hiraoka W, et al. Nucleoside analogs as a radiosensitizer modulating DNA repair, cell cycle checkpoints, and apoptosis. Nucleosides Nucleotides Nucleic Acids 2020;39:439-52. [Crossref] [PubMed]
- Gaillard H, García-Muse T, Aguilera A. Replication stress and cancer. Nat Rev Cancer 2015;15:276-89. [Crossref] [PubMed]
- Zhu H, Swami U, Preet R, et al. Harnessing DNA Replication Stress for Novel Cancer Therapy. Genes (Basel) 2020;11:990. [Crossref] [PubMed]
- Dillon MT, Good JS, Harrington KJ. Selective targeting of the G2/M cell cycle checkpoint to improve the therapeutic index of radiotherapy. Clin Oncol (R Coll Radiol) 2014;26:257-65. [Crossref] [PubMed]
- Vassilev LT. Cell cycle synchronization at the G2/M phase border by reversible inhibition of CDK1. Cell Cycle 2006;5:2555-6. [Crossref] [PubMed]
- Schmidt M, Rohe A, Platzer C, et al. Regulation of G2/M Transition by Inhibition of WEE1 and PKMYT1 Kinases. Molecules 2017;22:2045. [Crossref] [PubMed]
- Smith HL, Southgate H, Tweddle DA, et al. DNA damage checkpoint kinases in cancer. Expert Rev Mol Med 2020;22:e2. [Crossref] [PubMed]
- Vakili-Samiani S, Khanghah OJ, Gholipour E, et al. Cell cycle involvement in cancer therapy; WEE1 kinase, a potential target as therapeutic strategy. Mutat Res 2022;824:111776. [Crossref] [PubMed]
- Fu S, Wang Y, Keyomarsi K, et al. Strategic development of AZD1775, a Wee1 kinase inhibitor, for cancer therapy. Expert Opin Investig Drugs 2018;27:741-51. [Crossref] [PubMed]
- Taniguchi H, Caeser R, Chavan SS, et al. WEE1 inhibition enhances the antitumor immune response to PD-L1 blockade by the concomitant activation of STING and STAT1 pathways in SCLC. Cell Rep 2022;39:110814. [Crossref] [PubMed]
- Yang L, Shen C, Pettit CJ, et al. Wee1 Kinase Inhibitor AZD1775 Effectively Sensitizes Esophageal Cancer to Radiotherapy. Clin Cancer Res 2020;26:3740-50. [Crossref] [PubMed]
- Bukhari AB, Chan GK, Gamper AM. Targeting the DNA Damage Response for Cancer Therapy by Inhibiting the Kinase Wee1. Front Oncol 2022;12:828684. [Crossref] [PubMed]
- Sand A, Piacsek M, Donohoe DL, et al. WEE1 inhibitor, AZD1775, overcomes trastuzumab resistance by targeting cancer stem-like properties in HER2-positive breast cancer. Cancer Lett 2020;472:119-31. [Crossref] [PubMed]
- Chen X, Low KH, Alexander A, et al. Cyclin E Overexpression Sensitizes Triple-Negative Breast Cancer to Wee1 Kinase Inhibition. Clin Cancer Res 2018;24:6594-610. [Crossref] [PubMed]
- Iness AN, Felthousen J, Ananthapadmanabhan V, et al. The cell cycle regulatory DREAM complex is disrupted by high expression of oncogenic B-Myb. Oncogene 2019;38:1080-92. [Crossref] [PubMed]


