
Analysis of the role of glucose metabolism-related genes in dilated cardiomyopathy based on bioinformatics
Highlight box
Key findings
• PFKM, DLAT, PKLR, PGM2, LDHA, BPGM, ADH1A, and ADH1C are involved in the occurrence and development of cardiomyopathy.
What is known and what is new?
• The molecular mechanism of DCM has not been clearly elucidated, and the therapeutic targets of key genes in energy metabolism in DCM have not been extensively studied.
• PFKM, DLAT, PKLR, PGM2, LDHA, BPGM, ADH1A, and ADH1C may be involved in the occurrence and development of cardiomyopathy by regulating the Toll-like receptor signaling pathway.
What is the implication, and what should change now?
• In this study, the PFKM, DLAT, PKLR, PGM2, LDHA, BPGM, ADH1 and ADH1 genes were screened through bioinformatics technology and big data mining, and the functions and molecular mechanisms of these factors in DCM were analyzed. Our study suggests that these factors can be used as new targets to guide the diagnosis, treatment, and follow-up monitoring of patients with DCM.
Introduction
Dilated cardiomyopathy (DCM) is an idiopathic primary myocardial disease. It is characterized by the enlargement of the left or right ventricle or both, accompanied by impaired ventricular systolic function. It may or may not be accompanied by congestive heart failure (1). Despite extensive research efforts, there are still significant knowledge gaps and limitations in our understanding of DCM, particularly regarding its underlying mechanisms and the lack of clinical indicators for diagnosis, prevention, and monitoring (2). Currently, there is no specific therapy for DCM, and most patients are treated with beta blockers, ACE inhibitors, mineralocorticoid receptor antagonists, and SLG2 inhibitors to slow disease progression. However, the long-term prognosis remains poor with these treatments. At present, there is insufficient research into the pathogenesis of DCM, and there is also a lack of clinical indicators for the diagnosis, prevention, and monitoring of DCM. At present, there is insufficient research on the pathogenesis of DCM, and there is also a lack of clinical indicators for the diagnosis, prevention, and monitoring of DCM (3). Therefore, there is an urgent need to explore novel approaches to improve our understanding of DCM pathogenesis and identify potential molecular markers that can aid in diagnosis and guide effective treatment strategies.
The heart is a highly energy-consuming organ, which needs to consume a large amount of adenosine triphosphate (ATP) every day to complete cardiac pump function. Energy metabolism is mainly divided into glucose metabolism and fatty acid metabolism (4,5). Fatty acid oxidation accounts for about 70% of ATP produced by cardiac aerobic metabolism. However, the ability of the heart to synthesize and store fatty acids is limited, and its fatty acid supply mainly comes from plasma free fatty acids (FFA), lipoprotein lipase (LPL), and endogenous triacylglycerol (TAG). The glycogen reserve in the heart is low, and the glucose in glucose metabolism mainly comes from exogenous glucose. Glucose enters the cell via the insulin-dependent glucose transporter 1/4 (GLUT1/4) on the surface of cardiomyocytes. Theoretically, disorders of myocardial energy metabolism occur to varying degrees in all types of cardiac disease, but whether alterations in myocardial energy metabolism led to corresponding cardiac disease and whether disorders of fatty acid metabolism or glucose metabolism alone correspond to a specific type of cardiac disease need to be further explored. At present, study has shown that the key genes of glycolysis may be useful indicators to predict the prognosis of patients with liver cancer and guide clinical treatment (6).
Advances in bioinformatics technology and the availability of vast amounts of information on disease characteristics have given researchers a new framework for understanding the biology of diseases in the dimension of big data. Increasingly sophisticated machine learning algorithms, e.g., the non-negative matrix factorization (NMF) algorithm (7), least absolute shrinkage and selection operator (LASSO) regression algorithm (8), and the SVM method (9). These algorithms have been widely used in the screening, diagnosis, prognosis, and molecular target screening of a variety of diseases (10-12). The application of these bioinformatics techniques and machine algorithms can select the relevant factors in disease prevention, diagnosis, treatment, and monitoring using massive data sets, which greatly improves the efficiency of research on diseases (13). Previous study has reported the use of Gene Expression Omnibus (GEO) database combined with weighted gene coexpression network analysis (WGCNA) and the cell-type identification by estimating relative subsets of RNA transcripts (CIBERSORT) method to reveal regulatory relationships in DCM (14). Study has shown that miR-129-5p may regulate DCM by targeting ASPORIN gene through extracellular matrix (ECM) signaling pathway. Macrophage infiltration may participate in ECM remodeling and eventually lead to DCM (15).
In light of the aforementioned gaps in our understanding of DCM and the potential of glycolytic genes as molecular markers, this study aims to investigate the role of glycolysis in the occurrence and development of DCM and its underlying molecular regulatory mechanisms. Specifically, we will focus on eight signature glycolytic genes (PFKM, DLAT, PKLR, PGM2, LDHA, BPGM, ADH1A, and ADH1C) to explore their potential clinical significance in predicting DCM prognosis and guiding treatment strategies.
By employing a combination of bioinformatics technology and machine algorithms, our study seeks to contribute to a better understanding of DCM pathogenesis and provide insights into the development of effective diagnostic and therapeutic approaches. We present this article in accordance with the STREGA reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-906/rc).
Methods
Data sources
The GSE79962 (16) and GSE42955 (17) datasets were downloaded from the Gene Expression Omnibus (GEO) database. GSE42955 included 5 normal myocardial tissues and 12 DCM tissues. GSE79962 contained 11 normal myocardial tissues and 9 DCM tissues. The normal tissues and DCM samples of the 2 data sets were corrected in batches by the SVA program package (Bioconductor) and merged (18) to obtain a merged GEO-GSE79962 + GSE42955 (Merge) data set for subsequent analysis, including 16 normal myocardial tissues and 21 DCM tissues. A total of 23,306 genes were finally annotated in the GEO-Merge data set. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Differentially expressed gene (DEGs) screening
DEGs were screened in normal myocardial tissue and DCM tissue with the “limma” package in R software (The R Foundation for Statistical Computing), and the DEGs were defined with |log2FC (fold change)| >0.05 and P<0.05 as the screening criteria. The “ggplot2” software package in R was used to draw the volcano map of the DEGs of the GEO-GSE79962 + GSE42955 data set, and the “pheatmap” package of the R language was used to draw the heat map of the DEGs of the GEO-GSE79962 + GSE42955 data set.
Protein-protein interaction network and Gene Set Enrichment Analysis (GSEA)
GSEA software (version 4.2.2, Broad Institute, USA) was used to perform GSEA for all differential genes between DCM tissues and normal controls in the GEO-GSE79962 + GSE42955 data set. Biocarta Glycolysis Pathway, Go Glycolytic Process, Hallmark Glycolysis, Kegg Glycolysis Gluconeogenesis, and Reactome Glycolysis from the Molecular Signatures Database (MSigDB) were used as reference gene sets for Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis in visual analysis. The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) online database was used to predict the differentially expressed protein interactions, and the effective binding fraction was set to >0.7.
Calculation and screening of the characteristic glycolytic genes
In this study, the LASSO algorithm and SVM algorithm were combined to preselect the features of glycolytic DEGs, the receiver operating characteristic (ROC) curve of the model was drawn, and the area under the ROC curve (AUC) was calculated to evaluate the prediction efficiency of the model. The “Glmnet” package in R was used for LASSO regression analysis of the characteristic DEGs (19,20). LASSO combines feature selection and model building by adding penalty constraints to the algorithm. In the case of α=1, the appropriate λ value was selected through 10-fold cross validation. In the 10-fold cross validation, the gene combination with the smallest root-mean-square error and the highest accuracy was selected as the best gene combination (21). The recursive feature elimination (RFE) method was used to optimize and screen the feature set. The SVM method (9) was used to train the model, along with the linear fitting method. The 5-fold cross validation method (22) was used to randomly divide the samples into 5 parts: 4 parts were used as training sets to build SVM models, and 1 part was used for prediction sets to calculate accuracy. The above procedure was repeated 5 times until each forecast set was predicted only once as a forecast set.
Molecular typing of DCM samples
Non-negative matrix factorization (NMF) aims to decompose a nonnegative matrix into 2 nonnegative matrices (23), which has good interpretability and numerical results. This method has been widely used to classify gene expression profile data (24,25). In this study, the NMF molecular typing model was constructed using the “Consensus ClusterPlus” analysis package in R. The NMF hierarchical clustering was performed using the adjusted and unified data set, with the number of clusters k values ranging from 2 to 9 (26).
Predicting different percentages of immune cell infiltration in DCM samples
The CIBERSORT algorithm is a machine learning method based on linear support vector regression (SVR) and is highly robust to noise (27). The CIBERSORT deconvolution algorithm was used to analyze the infiltration of immune cells as well as the simulation calculation of the transcriptional feature matrix including 22 kinds of immune cells such as T cells, B cells, and natural killer (NK) cells (27). For accurate results, the number of simulations was set at 1,000, and the Kruskal-Wallis rank sum test was used for subsequent analysis of data at P<0.05.
To analyze the various types of immune cell correlations, Pearson correlation coefficients between different immune cells were calculated in the data of plausible samples screened by the CIBERSORT deconvolution algorithm on the basis of P<0.05, and the rank sum test was used to compare the differences between the two groups.
Statistical analysis
The data from GEO are merged using Perl software (Perl Foundation, USA). GSEA analysis with false discovery rate (FDR) <0.1, standardized enrichment fraction (normalized enrichment score, NES) >1 and P<0.05 for significant differences. Other statistical methods are described in the above materials and methods.
Results
Glycolysis involvement in the development of DCM
In order to clarify the role of glycolysis in DCM, the GEO-GSE79962 + GSE42955 data set was used to verify the enrichment degree of glycolytic genes in DCM. Our study has shown that, compared with normal myocardial tissues, glycolysis-related pathways were downregulated in DCM tissues (Figure 1A), and glycolysis gluconeogenesis was most significantly decreased (Figure 1B). These results suggested that glycolysis gluconeogenesis might be involved in the development of DCM.
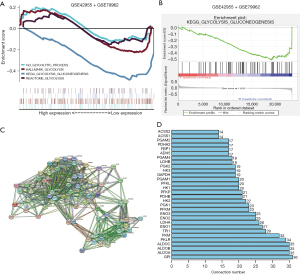
Subsequently, in order to further investigate the role of glycolysis in DCM, 62 genes related to glycolysis gluconeogenesis signaling pathway were extracted from the MSigDB website (Table S1). Firstly, the STRING website was used to construct a protein-protein interaction network for these 62 DEGs. The study showed that GPI, ALDOA, ALDOB, ALDOC, PKLR, PKM, TPI1, ENO1, LDHA, and ENO2 were the key node genes (Figure 1C,1D). Based on this, our conclusions suggest that glycolysis decreases in DCM, which may be related to the progression of DCM.
Identification and analysis of DEGs in glycolysis
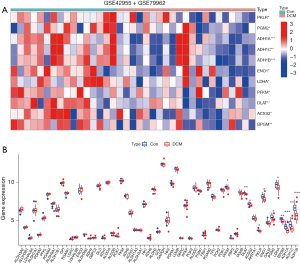
To clarify the differentially expressed genes of glycolysis in DCM, 62 genes related to Glycolysis Gluconeogenesis signaling pathway extracted from MSigDB website were used for differentially expressed gene analysis by applying GEO-Merge dataset, with |logFC| >0.5 and P<0.05 as the differentially expressed gene screening criteria. The study showed that 169 genes were differentially expressed in DCM compared with normal cardiac tissue. Through the intersection of genes related to the glycolysis gluconeogenesis signaling pathway, 11 glycolytic DEGs were obtained in this study, and all had a low expression in DCM. These were PFKM, DLAT, ACSS2, PKLR, ENO1, PGM2, LDHA, BPGM, ADH1A, ADH1C, and ADH1B genes (Figure 2A,2B). Based on this, 11 glycolytic-related genes that may play an important role in the development of DCM were screened by bioinformatics analysis.
Screening of DEGs in glycolysis
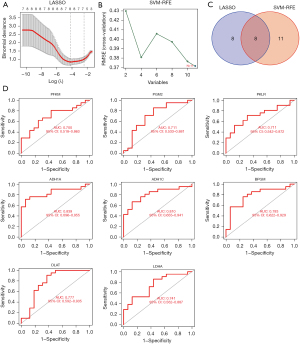
To further focus on factors with clinical translational potential, we performed signature gene analysis on these 11 glycolytic-related genes. We combined the LASSO algorithm and SVM algorithm to analyze these 11 glycolytic-related genes. Results showed that the LASSO algorithm obtained 8 candidate feature genes (PFKM, DLAT, PKLR, PGM2, LDHA, BPGM, ADH1A, and ADH1C) (Figure 3A). The SVM algorithm obtained 11 candidate feature genes (PFKM, DLAT, ACSS2, PKLR, ENO1, PGM2, LDHA, BPGM, ADH1A, ADH1C, and ADH1B genes) (Figure 3B). The intersection of the 2 algorithms was conducted to obtain 8 candidate feature genes (Figure 3C), which were the PFKM, DLAT, PKLR, PGM2, LDHA, BPGM, ADH1A, and ADH1C genes. Further AUC analysis of these 8 alternative feature factors indicated AUC values for PFKM, DLAT, PKLR, PGM2, LDHA, BPGM, ADH1A, and ADH1C of 0.700, 0.777, 0.711, 0.711, 0.741, 0.783, 0.839, and 0.810, respectively (Figure 3D). Based on this, we focused on 8 genes from the 11 glycolytic genes.
Establishment of molecular classification of DCM
Traditional disease diagnosis relies on pathology (28); however, pathology has difficulty in discerning the biological nature of tumors. Different diseases show different biological characteristics, which may be related to the different molecular composition and expression of the lesions. New disease typing methods based on molecular typing can provide global characteristics of the disease gene level, greatly deepen our understanding of molecular pathological information, and assume an important role in clinical practice. Therefore, in order to verify whether the 8 feature genes obtained by the above-described screening had clinical transformation potential, we used these 8 feature genes combined with the GEO-Merge dataset to study the molecular typing of DCM.
According to the enrichment scores of 8 genes, we used K-means consistent clustering to cluster 21 samples of DCM. However, stable clustering results could not be obtained when k=2−9 (Figure 4A-4D); that is, samples of DCM could not be classified based on these 8 characteristic genes, suggesting that these 8 genes do not have a clinical staging effect, and their mechanism of action needs to be further studied.

Effect of the LDHA and ADH1C on the immune microenvironment of DCM
We used the GEO-Merge dataset to compare immune cell infiltration in normal and DCM tissues. Compared with normal myocardial tissues, regulatory T cells (Tregs), activated dendritic cells, and activated mast cells were downregulated in DCM tissues (Figure 5A, red), while M0 macrophages were upregulated in DCM tissues (Figure 5A, blue). These results suggest that activated dendritic cells, activated mast cells, and M0 macrophages may be involved in the immune injury of DCM.
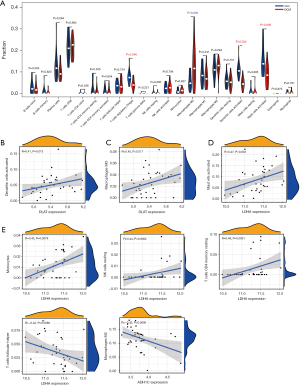
Next, the 8 characteristic glycolytic genes were compared with the immune cells (Tregs, activated dendritic cells, activated mast cells, and M0 macrophages) for correlation analysis. The results showed that DLAT was moderately positively correlated with activated dendritic cells (R=0.41; Figure 5B), while M0 macrophages were moderately positively correlated with DLAT (R=0.40; Figure 5C). There was a moderate positive correlation between LDHA and activated mast cells (R=0.47; Figure 5D). Moreover, our results also suggested that the LDHA and ADH1C genes were correlated with the presence of other immune cells to varying degrees (Figure 5E). These data suggest that DLAT and LDHA may influence the progression of DCM through their expression correlation with activated dendritic cells, activated mast cells, and M0 macrophages. However, the association here does not imply causality, and further research is required to confirm the role of DLAT and LDHA in regulating these immune cell subpopulations.
BPGM, DLAT, PGM2, ADH1A, ADH1C, LDHA, and PFKM genes affected m6A methylation in DCM
First, we used the GEO-Merge data set to compare the differential expression of m6A methylation-related genes between normal myocardial tissues and DCM tissues. Our study showed that METTL3, ZC3H13, YTHDC1, HNRNPC, RBMX, and ALKBH5 were differentially expressed in DCM tissues compared with normal myocardial tissues (Figure 6A). The expressions of METTL3, ZC3H13, YTHDC1, and HNRNPC genes were significantly decreased in DCM, while the expressions of RBMX and ALKBH5 were significantly increased in DCM (Figure 6B).

Further correlation analysis verified that BPGM, DLAT, and PGM2 were moderately negatively correlated with the ZC3H13 gene (R<−0.4) (Figure 6C); the LDHA and HNRNPC genes were moderately negatively correlated (R<−0.4) (Figure 6C); ADH1C was moderately negatively correlated with the METTL3 gene (R<−0.4) (Figure 6C). Similarly, BPGM was moderately positively correlated with the ALKBH5 gene (R>0.4) (Figure 6D), while BPGM and PFKM were moderately positively correlated with the RBMX gene (R>0.4) (Figure 6D). These results suggest that the BPGM, DLAT, PGM2, ADH1A, ADH1C, LDHA, and PFKM genes have interactions with m6A methylation-related genes, and such interactions may indirectly influence the m6A methylation levels in DCM, thereby affecting the progression of DCM. However, further research is needed to investigate the precise relationship between these genes and m6A methylation.
Identification and verification of the regulatory mechanisms of 8 characteristic glycolytic genes
We used the GEO-Merge data set to study the regulatory mechanism of 8 characteristic glycolytic genes in DCM. First, we performed GSEA analysis on these 8 genes, which were revealed to be enriched in 366 pathways (Table S2). Furthermore, after the enrichment pathways of each gene were intersected, we found Toll-like receptor signaling pathway, complement and coagulation cascades, cytokine receptor interaction, FC gamma R-Mediated phagocytosis, hematopoietic cell lineage, and systemic lupus erythematosus to be the common pathways of the 8 characteristic genes (Figure 7A, down). This suggests that these signaling pathways may be involved in the development of cardiomyopathy, and targeting key factors in these pathways may slow down the progression of DCM.

Next, we focused on the Toll-like receptor signaling pathway, because there is little research on the relationship between the Toll-like receptor signaling pathway and DCM, and it may thus be a therapeutic target for DCM. Therefore, in order to find a clear target and regulatory mechanism, we analyzed the effects of the PFKM, DLAT, PKLR, PGM2, LDHA, BPGM, ADH1A, and ADH1C genes on TLR1, TLR2, TLR3, TLR4, TLR5, TLR6, TLR7 and TLR8. The results showed that TLR1 and TLR8 were each correlated with 5 glycolytic genes (Figure 7B); TLR2, TLR4, and TLR6 were each correlated with 4 glycolytic characteristic genes (Figure 7B); and TLR3, TLR5, and TLR7 were each correlated with 2 glycolytic characteristic genes (Figure 7B). Based on the above, our data revealed that the PFKM, DLAT, PKLR, PGM2, LDHA, BPGM, ADH1A, and ADH1C genes may be involved in the development of DCM by regulating the Toll-like receptor signaling pathway.
Discussion
The treatment of DCM primarily involves comprehensive management, which includes the combined application of medication, surgical interventions, cardiac rehabilitation, lifestyle management, and supportive therapies (29). Although this can improve the patient’s condition, there are still problems such as poor prognosis, which may be caused by its onset. Due to insufficient molecular mechanism research and a lack of molecular markers for clinical diagnosis, treatment, and prognosis, effective monitoring and intervention in the early stage of the disease remains elusive. Therefore, it is extremely important to identify the early diagnostic biomarkers that can help detect and possibly prevent the occurrence of DCM (30,31).
Cardiac diseases are closely related to energy metabolism, and changes in myocardial energy metabolism occur in various cardiac diseases, such as myocardial hypertrophy (32,33), ischemic heart disease (34,35), diabetes mellitus (36), and heart failure (37). Theoretically speaking, various degrees of myocardial energy metabolism disorders may occur in any type of heart disease, but whether the changes in myocardial energy metabolism correspond to a given type of heart disease still needs to be further explored. Therefore, in-depth study of the relationship between changes in myocardial energy metabolism and myocardial diseases is conducive to providing new therapeutic concepts for myocardial diseases from the perspective of energy metabolism.
At present, there is literature supporting the notion that serum metabolites can be used as biomarkers in DCM. In one study, researchers found that metabolites including lactate, succinate, and malate were elevated in patients with DCM (38). In another study, researchers measured 149 metabolites in 273 plasma and urine samples from patients with DCM at different disease stages and found that acylcarnitines, sialic acids, and glutamate were associated with DCM severity (39). However, human DCM metabolomics study have only been performed on biological fluids, such as serum, plasma, or urine, in part because blood is noninvasive and readily available, but the local biochemical information of DCM cannot be easily ascertained. Therefore, tissue analysis from DCM lesions may be the most powerful method to study the pathogenesis of DCM, as this can obtain clear biochemical information about the disease mechanism (40).
Based on this, this study took glucose metabolism and the DCM gene chip as the entry point. We identified the low expression of glycolysis in DCM tissues through GSEA, which suggested that the glycolysis process was inhibited in DCM tissues. Then, using the GEOGSE79962 + GSE42955 data set, we found 11 differentially expressed glycolytic-related genes (all genes were downregulated) in DCM tissues, indicating potential roles of these genes in the pathogenesis of DCM. However, these findings need to be validated through further experimental research and longitudinal data.
Based on this, this study took glucose metabolism and the DCM gene chip as the entry point. We identified the low expression of glycolysis in DCM tissues through GSEA, which suggested that the glycolysis process was inhibited in DCM tissues. Then, using the GEOGSE79962 + GSE42955 dataset, we found 11 differentially expressed glycolytic-related genes (all genes were downregulated) DCM tissues, suggesting that these genes may be involved in the regulation of the progression of DCM and thus may be used as new targets to guide clinical diagnosis, treatment, and prognosis.
In order to screen new targets of DCM, the LASSO algorithm and SVM algorithm were used to analyze these 11 glycolytic-related genes. This had the advantage of increasing the efficiency of target screening. Through the machine algorithm, 8 characteristic glycolytic genes with high correlation with DCM and differential expression in DCM were finally screened in this study. Moreover, further analysis of the diagnostic effect of these 8 characteristic genes also reached above 0.7, indicating that they have a good clinical translation potential.
In the next step, in order to have a more intuitive understanding of the clinical significance of these 8 characteristic factors and to analyze how these 8 characteristic factors affect DCM, we conducted a series of analyses.
First, we used these 8 characteristic factors to perform molecular typing of DCM (NMF algorithm). However, this study showed that DCM could not be typed molecularly based on the expression levels of these 8 genes, which indicated the limitations of taking the 8 characteristic factors as a whole to conduct a diagnosis of DCM.
Second, our study analyzed the immune infiltration pattern of DCM. By utilizing bioinformatics tools such as CIBERSORT, we assessed the relative proportions of different types of immune cells in the myocardial tissue of DCM patients (15). We found that immune factors may be involved in the occurrence and development of DCM and DLAT and LDHA may affect the course of DCM by regulating the distribution of immune cells.
By analyzing the immune infiltration characteristics of DCM, we can gain a deeper understanding of the disease’s pathogenesis and progression. This information may help guide future research and development of immunotherapeutic strategies for DCM. For instance, modulating immune system activity or designing drugs targeting specific immune cell types may bring new breakthroughs in the treatment of DCM.
Third, studies have shown that m6A methylation plays an important regulatory role in heart failure (41,42), myocardial hypertrophy (43,44), atherosclerosis (45), ischemic cardiomyopathy (46,47), and other cardiovascular diseases. In this study, we applied the GEO-Merge dataset to compare the differential expression of m6A methylation-related genes in normal and DCM tissues. It was found that METTL3, ZC3H13, YTHDC1, HNRNPC and RBMX genes were significantly decreased in expression in DCM; while RBMX and ALKBH5 were significantly increased in DCM, suggesting that they may be involved in the progression of DCM. Next, in this study, the correlation between the 8 characteristic factors and m6A methylation-related genes was analyzed. The conclusion of our study showed that 7 glycolytic characteristic factors were correlated with 6 m6A methylation-related genes, which also indicated that these characteristic factors could affect the expression of m6A methylation in local tissues of DCM.
In order to explain the regulatory mechanism of the 8 characteristic glycolytic genes, enrichment analysis of the selected 8 key genes was carried out. GSEA analysis method avoids the problem of threshold setting in the traditional enrichment analysis method, and the whole genome data are included in enrichment analysis. In this study, 8 key genes were enriched in 6 common pathways. Furthermore, we focused on the Toll-like receptor signaling pathway because its mechanism of action in DCM remains poorly explained. Through correlation analysis with key factors of Toll-like receptor signaling pathway, our data revealed that the PFKM, DLAT, PKLR, PGM2, LDHA, BPGM, ADH1A, and ADH1C genes may be involved in the development of cardiomyopathy by regulating the Toll-like receptor signaling pathway.
The results of this study have the potential to aid in the development of therapeutic strategies against DCM. Firstly, we identified core genes closely associated with DCM, which may serve as potential therapeutic targets. By gaining a deeper understanding of the functions and interactions of these genes, we can explore drugs or interventions that may modulate these genes, thereby providing new strategies for the treatment of DCM.
Additionally, this study lays the foundation for future clinical research. For instance, in the development of new drugs targeting DCM, the identified core genes can serve as biomarkers for evaluating drug efficacy. By monitoring changes in the expression of these core genes before and after drug treatment, we can assess a patient’s response to therapy, enabling personalized treatment.
Conclusions
In this study, we identified 8 signature glycolytic genes in DCM and elucidated their functions and mechanisms. The genes PFKM, DLAT, PKLR, PGM2, LDHA, BPGM, ADH1A, and ADH1C appear to be novel biomarkers of DCM. Future studies are needed to elucidate the biological processes involved in the regulation of these glycolytic signature genes and their respective roles in the initiation and progression of DCM.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-906/rc
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-906/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-906/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Merlo M, Cannatà A, Gobbo M, et al. Evolving concepts in dilated cardiomyopathy. Eur J Heart Fail 2018;20:228-39. [Crossref] [PubMed]
- Hershberger RE, Morales A, Siegfried JD. Clinical and genetic issues in dilated cardiomyopathy: a review for genetics professionals. Genet Med 2010;12:655-67. [Crossref] [PubMed]
- Huang Y, Zhang J, Xu D, et al. SIRT6-specific inhibitor OSS-128167 exacerbates diabetic cardiomyopathy by aggravating inflammation and oxidative stress. Mol Med Rep 2021;23:367. [Crossref] [PubMed]
- Meng J, Ma N, Liu H, et al. Untargeted and targeted metabolomics profiling reveals the underlying pathogenesis and abnormal arachidonic acid metabolism in laying hens with fatty liver hemorrhagic syndrome. Poult Sci 2021;100:101320. [Crossref] [PubMed]
- Hayasaka K. Metabolic basis and treatment of citrin deficiency. J Inherit Metab Dis 2021;44:110-7. [Crossref] [PubMed]
- Chen Q, Bao L, Huang Y, et al. Clinical significance and immunogenomic landscape analysis of glycolysis-associated prognostic model to guide clinical therapy in hepatocellular carcinoma. J Gastrointest Oncol 2022;13:1351-66. [Crossref] [PubMed]
- Gaujoux R, Seoighe C. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics 2010;11:367. [Crossref] [PubMed]
- Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med 1997;16:385-95. [Crossref] [PubMed]
- Ma S, Lv M, Deng F, et al. Predicting the ecotoxicity of ionic liquids towards Vibrio fischeri using genetic function approximation and least squares support vector machine. J Hazard Mater 2015;283:591-8. [Crossref] [PubMed]
- Rau CS, Wu SC, Chuang JF, et al. Machine Learning Models of Survival Prediction in Trauma Patients. J Clin Med 2019;8:799. [Crossref] [PubMed]
- Lee HC, Yoon HK, Nam K, et al. Derivation and Validation of Machine Learning Approaches to Predict Acute Kidney Injury after Cardiac Surgery. J Clin Med 2018;7:322. [Crossref] [PubMed]
- Szlosek DA, Ferrett J. Using Machine Learning and Natural Language Processing Algorithms to Automate the Evaluation of Clinical Decision Support in Electronic Medical Record Systems. EGEMS (Wash DC) 2016;4:1222. [Crossref] [PubMed]
- Karakulak T, Rifaioglu AS, Rodrigues JPGLM, et al. Predicting the Specificity- Determining Positions of Receptor Tyrosine Kinase Axl. Front Mol Biosci 2021;8:658906. [Crossref] [PubMed]
- Zhu Y, Yang X, Zu Y. Integrated analysis of WGCNA and machine learning identified diagnostic biomarkers in dilated cardiomyopathy with heart failure. Front Cell Dev Biol 2022;10:1089915. [Crossref] [PubMed]
- Yang Y, Liu P, Teng R, et al. Integrative bioinformatics analysis of potential therapeutic targets and immune infiltration characteristics in dilated cardiomyopathy. Ann Transl Med 2022;10:348. [Crossref] [PubMed]
- Zhang K, Wu M, Qin X, et al. Asporin is a Potential Promising Biomarker for Common Heart Failure. DNA Cell Biol 2021;40:303-15. [Crossref] [PubMed]
- Liu C, Chen S, Zhang H, et al. Bioinformatic analysis for potential biological processes and key targets of heart failure-related stroke. J Zhejiang Univ Sci B 2021;22:718-32. [Crossref] [PubMed]
- Hivert MF, Cardenas A, Allard C, et al. Interplay of Placental DNA Methylation and Maternal Insulin Sensitivity in Pregnancy. Diabetes 2020;69:484-92. [Crossref] [PubMed]
- Liu Z, Mi M, Li X, et al. A lncRNA prognostic signature associated with immune infiltration and tumour mutation burden in breast cancer. J Cell Mol Med 2020;24:12444-56. [Crossref] [PubMed]
- Kang J, Choi YJ, Kim IK, et al. LASSO-Based Machine Learning Algorithm for Prediction of Lymph Node Metastasis in T1 Colorectal Cancer. Cancer Res Treat 2021;53:773-83. [Crossref] [PubMed]
- Li M, Li X, Guo Y, et al. Development and assessment of an individualized nomogram to predict colorectal cancer liver metastases. Quant Imaging Med Surg 2020;10:397-414. [Crossref] [PubMed]
- Mou W, Liu Z, Luo Y, et al. Development and cross-validation of prognostic models to assess the treatment effect of cisplatin/pemetrexed chemotherapy in lung adenocarcinoma patients. Med Oncol 2014;31:59. [Crossref] [PubMed]
- Lee DD, Seung HS. Learning the parts of objects by non-negative matrix factorization. Nature 1999;401:788-91. [Crossref] [PubMed]
- Bayar B, Bouaynaya N, Shterenberg R. Probabilistic non-negative matrix factorization: theory and application to microarray data analysis. J Bioinform Comput Biol 2014;12:1450001. [Crossref] [PubMed]
- Ching T, Peplowska K, Huang S, et al. Pan-Cancer Analyses Reveal Long Intergenic Non-Coding RNAs Relevant to Tumor Diagnosis, Subtyping and Prognosis. EBioMedicine 2016;7:62-72. [Crossref] [PubMed]
- Lancichinetti A, Fortunato S. Consensus clustering in complex networks. Sci Rep 2012;2:336. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. [Crossref] [PubMed]
- Han XY, Wang YY, Wei HQ, et al. Multifocal neuroendocrine cell hyperplasia accompanied by tumorlet formation and pulmonary sclerosing pneumocytoma: A case report. World J Clin Cases 2020;8:3583-90. [Crossref] [PubMed]
- Orphanou N, Papatheodorou E, Anastasakis A. Dilated cardiomyopathy in the era of precision medicine: latest concepts and developments. Heart Fail Rev 2022;27:1173-91. [Crossref] [PubMed]
- Favalli V, Serio A, Grasso M, et al. Genetic causes of dilated cardiomyopathy. Heart 2016;102:2004-14. [Crossref] [PubMed]
- Dec GW, Fuster V. Idiopathic dilated cardiomyopathy. N Engl J Med 1994;331:1564-75. [Crossref] [PubMed]
- Seymour AM, Giles L, Ball V, et al. In vivo assessment of cardiac metabolism and function in the abdominal aortic banding model of compensated cardiac hypertrophy. Cardiovasc Res 2015;106:249-60. [Crossref] [PubMed]
- Pereira RO, Wende AR, Olsen C, et al. GLUT1 deficiency in cardiomyocytes does not accelerate the transition from compensated hypertrophy to heart failure. J Mol Cell Cardiol 2014;72:95-103. [Crossref] [PubMed]
- Bayeva M, Gheorghiade M, Ardehali H. Mitochondria as a therapeutic target in heart failure. J Am Coll Cardiol 2013;61:599-610. [Crossref] [PubMed]
- Roselló-Lletí E, Tarazón E, Barderas MG, et al. ATP synthase subunit alpha and LV mass in ischaemic human hearts. J Cell Mol Med 2015;19:442-51. [Crossref] [PubMed]
- Stanley WC, Recchia FA, Lopaschuk GD. Myocardial substrate metabolism in the normal and failing heart. Physiol Rev 2005;85:1093-129. [Crossref] [PubMed]
- Aksentijević D, McAndrew DJ, Karlstädt A, et al. Cardiac dysfunction and peri-weaning mortality in malonyl-coenzyme A decarboxylase (MCD) knockout mice as a consequence of restricting substrate plasticity. J Mol Cell Cardiol 2014;75:76-87. [Crossref] [PubMed]
- Haas J, Frese KS, Sedaghat-Hamedani F, et al. Energy Metabolites as Biomarkers in Ischemic and Dilated Cardiomyopathy. Int J Mol Sci 2021;22:1999. [Crossref] [PubMed]
- Verdonschot JAJ, Wang P, Van Bilsen M, et al. Metabolic Profiling Associates with Disease Severity in Nonischemic Dilated Cardiomyopathy. J Card Fail 2020;26:212-22. [Crossref] [PubMed]
- Johnson CH, Ivanisevic J, Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol 2016;17:451-9. [Crossref] [PubMed]
- Berulava T, Buchholz E, Elerdashvili V, et al. Changes in m6A RNA methylation contribute to heart failure progression by modulating translation. Eur J Heart Fail 2020;22:54-66. [Crossref] [PubMed]
- Zhang B, Xu Y, Cui X, et al. Alteration of m6A RNA Methylation in Heart Failure With Preserved Ejection Fraction. Front Cardiovasc Med 2021;8:647806. [Crossref] [PubMed]
- Qian B, Wang P, Zhang D, et al. m6A modification promotes miR-133a repression during cardiac development and hypertrophy via IGF2BP2. Cell Death Discov 2021;7:157. [Crossref] [PubMed]
- Zhou W, Wang C, Chang J, et al. RNA Methylations in Cardiovascular Diseases, Molecular Structure, Biological Functions and Regulatory Roles in Cardiovascular Diseases. Front Pharmacol 2021;12:722728. [Crossref] [PubMed]
- Zhang BY, Han L, Tang YF, et al. METTL14 regulates M6A methylation-modified primary miR-19a to promote cardiovascular endothelial cell proliferation and invasion. Eur Rev Med Pharmacol Sci 2020;24:7015-23. [PubMed]
- Song H, Feng X, Zhang H, et al. METTL3 and ALKBH5 oppositely regulate m(6)A modification of TFEB mRNA, which dictates the fate of hypoxia/reoxygenation-treated cardiomyocytes. Autophagy 2019;15:1419-37. [Crossref] [PubMed]
- Chen X, Wang J, Tahir M, et al. Current insights into the implications of m6A RNA methylation and autophagy interaction in human diseases. Cell Biosci 2021;11:147. [Crossref] [PubMed]
(English Language Editor: J. Gray)


