
K. pneumoniae and M. smegmatis infect epithelial cells via different strategies
Highlight box
Key findings
• K. pneumoniae and M. smegmatis could stimulate the secretion of specific cytokines and chemokines through transcription regulations in lung epithelial cell A549. The pathogen-epithelial cell interaction and the crosstalk among immune cells were thus changed.
What is known and what is new?
• M. smegmatis has stronger adhesion and intracellular retention ability, while K. pneumoniae has a higher intracellular replication ability.
• There existed significant correlations between K. pneumoniae infection and higher levels of IL-15, IL-1Ra, FGF basic, and G-CSF. At the same time, K. pneumoniae infection also changed the expression of F-actin in epithelial cells.
What are the implications, and what should change now?
• Different pathogens damaged the host through diverse strategies. K. pneumoniae was more likely to form acute infection compared with M. smegmatis.
Introduction
Bacterial pneumonia is a common severe disease. It can develop into sepsis and is difficult to eradicate when caused by multidrug-resistant strains. Klebsiella pneumoniae (K. pneumoniae) is a Gram-negative bacterium belonging to the Enterobacteriaceae family, capable of causing a variety of infections, such as pneumonia, urinary tract infections, wound infections, and bacteremia. K. pneumoniae, the third leading cause of nosocomial infections, poses a significant public health risk (1), especially in the population of immunosuppressed patients. Further compounding this concern, K. pneumoniae promptly develops antibiotic resistance, making it more difficult to select the appropriate antibiotic treatment (2,3). Resistance to carbapenems appears to have the greatest influence on the effectiveness of treatment. K. pneumoniae has vital virulence factors, such as a capsule, lipopolysaccharides, siderophores, and adhesins, which are necessary for the organism’s mechanisms of colonization, adhesion, invasion, and infection progression (4,5). Additionally, heat-stable enterotoxins, tyrosine kinase, heat-labile exotoxins, and hemolysins all contribute to the pathogenicity (6,7). A research set up K. pneumoniae cell-free DNA (cfDNA) detection system, but the clinical application value still needs further study (8). Despite extensive research over the past few decades, the pathophysiology of K. pneumoniae, especially its internalization of alveolar cells, remains poorly understood. It may be possible to enhance the existing therapeutic approaches and vaccine development with a better understanding of the innate immune response to K. pneumoniae infection.
Like K. pneumoniae, M. smegmatis represents an important class of opportunistic pathogens. Additionally, the prevalence of non-tuberculous mycobacterial (NTM) lung infections, including cystic fibrosis (CF), Human immunodeficiency virus (HIV) infection, and chronic respiratory diseases like bronchiectasis and chronic obstructive pulmonary diseases (COPDs), has significantly increased recently, especially in immunocompromised patients. Due to their high morbidity and death rates, mycobacterial infections were a significant medical concern during the acquired immunodeficiency syndrome (AIDS) pandemic (9). NTM includes over 150 species, excluding Mycobacterium tuberculosis (MTB) or M. leprae (10). Most of these species are aerobic, Gram-positive, non-motile organisms with hard and thick cell walls (11,12). As environmental bacteria, they are ubiquitous in soil and natural water sources, and most of them are opportunistic pathogens (9). A review indicated that 1.5% of NTM-induced lung infections were caused by the M. smegmatis group. In addition, such a high distribution is consistent with the pattern seen in South Korea and India (13). Pneumonia, cellulitis, localized abscesses, and osteomyelitis following trauma can all result from community-acquired M. smegmatis group infections (14,15). Various wound and soft tissue infections are examples of healthcare-associated infections (15,16). Disseminated infection has also been noted (7). With the history of antibiotic usage, this bacterium has evolved some mechanisms for resistance against the majority of drugs (17). However, the mechanisms underlying the rising incidence of M. smegmatis infection in people remain ill-defined.
A host can become infected by pathogenic microorganisms through various mechanisms. While avoiding host defenses, respiratory infections are breathed into the lungs, where they adhere to and traverse the respiratory mucosa. The interaction with host proteins and the modification of host cell signaling are frequent mediators of the capacity of bacteria to adhere to and infiltrate the mucosal epithelium. In response to pathogen invasion, cytokines and chemokines are essential chemicals that play a role in practically every aspect of immunity and inflammation (18,19). It has been determined that interleukin (IL)-6 increases neutrophil death to improve the survival of pneumonia sepsis brought on by K. pneumoniae (20). In mice, it has been shown that chemokine ligand 1 (CXCL1) can increase resistance to K. pneumonia (21). MTB can not only invade type II alveolar epithelial cells (AECs) but also replicate in cells (22,23). In fact, as intracellular pathogens, both K. pneumoniae and M. smegmatis can effectively “escape” the effect of antibacterial drugs, which highlights the need for a better understanding of biology in terms of the complex interaction between bacterial pathogens and their host cells.
Numerous pathogens can enter the body through the lung. The pulmonary alveolar space is the largest region of interaction between the body and the outside world. Type II AECs are cuboidal cells that function as stem cells for the alveolar epithelium and create pulmonary surfactant. The vast majority of the alveolar surface is covered with Type I AECs. Bacterial pathogens colonize host tissues by first adhering to epithelial cells, followed by entering cells through internalization (24). Due to this phenomenon, bacteria are shielded from the host’s defensive systems and the bactericidal effects of antibiotics, which hardly ever reach epithelial cells. Recent data suggest that AECs may be crucial in the control of inflammatory and immunological reactions in the lung (25). Particularly, type II AECs (AT-II) cells have diverse functions to protect against pathogens by sensing pathogens via toll-like receptor (TLR) stimulation (26,27), the secretion of anti-microbial peptides (28), and activating and deactivating inflammation by regulating cytokines and chemokines (29). It has been demonstrated that dangerous bacteria, including M. smegmatis and K. pneumoniae, may internalize into AT-II cells and survive these anti-microbial defenses. This even enables the bacteria to pass the alveolar barrier and enter the circulation.
Owing to the lung’s extensive set of defenses against this ongoing exposure to microbial pathogens, pneumonia is an uncommon side effect of recurrent low-level bacterial infiltration into the peripheral lung (25). According to experimental and epidemiological studies, a pathogen’s immune response can affect a second, unrelated pathogen that is being controlled by the host, changing the course of co-infection. In the face of complex saturation in the lung, it is very important to clarify the immunological mechanisms of different pathogens and obtain comparable data for the effective prevention and treatment of pulmonary bacterial infections, including the development of vaccines or immunotherapy.
At present, there have been several studies exploring the pathogenesis of bacterial pathogens using cell lines, animal models, or clinical samples. In the majority of these studies, only a single pathogen was chosen as the research object. There are some studies on the co-infection of viruses and pathogenic bacteria, but unfortunately, to our knowledge, there are no reports comparing K. pneumoniae and M. smegmatis under the same experimental conditions. Comparing the outcomes of several studies can be challenging due to variations in the cell, animal, and study designs. As such, no consistent conclusions can be drawn from them. In the present study, we performed a head-to-head comparison of the virulence and infection mechanism of K. pneumoniae and M. smegmatis, which represent Gram-negative/positive respiratory pathogens, respectively, in lung epithelial cell models. Investigating the function of lung epithelial cells in bacterial pneumonia and the determination of whether bacterial interactions with epithelial cells contribute to the pathogenesis of K. pneumoniae and M. smegmatis pneumonia were additional objectives of this study. We present this article in accordance with the MDAR reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-493/rc).
Methods
Strains and growth media
K. pneumoniae was cultured in Luria-Bertani broth (LB) at 37 ℃ and 180 rpm under aerobic conditions. For cell enumeration experiments, K. pneumoniae was plated on trypticase soy agar plates supplemented with 5% sheep’s blood (TSA). Serial 10-fold dilutions in phosphate buffered saline solution (PBS) were plated on TSA agar plates and incubated for 18 hours at 37 ℃, with each dilution plated in triplicate.
M. smegmatis (MC2 155) was grown in Middlebrook 7H9 broth (DIFCO, USA) supplemented with 10% albumin-dextrose-catalase (ADC) (BD, USA), 0.2% glycerol, and 0.05% Tween 80. The cultures were grown at 37 ℃ and 220 rpm under aerobic conditions. For cell enumeration experiments, M. smegmatis was plated on Middlebrook 7H10 agar (DIFCO, USA) containing 10% oleic acid-albumin-dextrose-catalase (OADC). Serial 10-fold dilutions in PBS were prepared and added on 7H10 agar plates then incubated for 24 hours at 37 ℃, with each dilution plated in triplicate.
The colonies were calculated and the number of surviving bacteria was exhibited in log10 units.
Growth curve in vitro
To determine the phenotypic growth characteristics of K. pneumoniae and M. smegmatis in vitro, K. pneumoniae and M. smegmatis strains were subcultured in liquid medium three times [optical density (OD) at 600 nm =0.8]. For the growth curves experiment, a 100 mL triangular flask containing 40 mL of LB broth was inoculated with K. pneumoniae to have an initial OD =0.05, and a 100 mL triangular flask containing 40 mL of 7H9 broth with ADC was inoculated with M. smegmatis to have an initial OD =0.05. The growth curves were monitored by the OD at 600 nm at 3 h intervals. The strains were then cultured in a 37 ℃ shaker under the same culture conditions. The experiments were performed in triplicate.
Scanning electron microscopy
Scanning electron microscopy was used to examine the minor changes in the bacterial morphology of the population. K. pneumoniae or M. smegmatis cultures (5 mL each) grown to mid-log phase were centrifuged at 8,000 rpm for 10 min and the cells were collected. The cells were rinsed with 0.1 M phosphate buffer (PB) two times and then fixed with 2.5% glutaraldehyde in 0.1 M PB (2 h, 4 ℃). The pellet was then re-suspended in 0.1 mL PBS. After centrifugation, the cells were fixed with 1% osmic acid (1.5 h, 4 ℃) and then washed with 0.1 M PBS (pH 7.2). The cells were dehydrated in a graded series of ethanol (10% to 100%) for 5 min at each concentration. Afterwards, the samples were microscopic examined by a SU5000 scanning electron microscope (Philips, USA) after being sputter coated with gold under a vacuum.
Cultures of epithelial cell line A549
The A549 human alveolar type II epithelial cell line was stored in our lab. Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco, Foster City, CA, USA) supplemented with 10% fatal bovine serum (FBS) (Gibco), 100 Unit/mL penicillin, and 100 µg/mL streptomycin (Gibco) was used to culture the A549 cells. The cells were preserved at 37 ℃ in a humidified atmosphere containing 5% carbon dioxide (CO2).
Analysis of the intracellular growth of K. pneumoniae or M. smegmatis
Infection of the human lung epithelial cells and enumeration of intracellular K. pneumoniae or M. smegmatis were executed as introduced by Vadivelu et al. (30). Briefly, confluent monolayers of A549 cells were cultured in 24-well tissue culture plates at a density of 105 cells per well in RPMI 1640 medium (10% FBS) for 24 h. A549 cells were infected with K. pneumoniae at a multiplicity of infection (MOI) of 1 for 3 h at 37 ℃ in 5% CO2 to allow for invasion. Extracellular bacteria were eliminated by incubating the tissue culture with 200 g/mL gentamycin for 2 h after two sterile PBS (pH 7.0) washes in the wells. The cell monolayers were then lysed with 1% Triton X-100 after being rinsed twice with PBS. The proportion of invasion was estimated after the lysate was serially diluted and plated onto TSA agar to count the bacteria.
For the purpose of determining K. pneumoniae’s capacity for intracellular replication, a similar procedure was used. Briefly, the cell monolayers were washed two times with PBS and incubated at 37 ℃ in 5% CO2 in fresh media with 20 µg/mL gentamycin. The infected monolayers (three wells per strain) were washed three times with fresh media and then lysed using 0.1 mL of 1% Triton X-100, and the intracellular bacteria released were enumerated by plating the serial dilution of the lysate on TSA agar. Two independent experiments were performed.
For M. smegmatis, the assay was performed as above with slight modifications: MOI of 25 for 3 h, and M. smegmatis was plated on Middlebrook 7H10 agar supplemented with OADC for cell enumeration.
Cell viability test with Cell Counting Kit-8 (CCK-8)
CCK-8 (Tongren Chemical Company, China), which is a rapid, highly sensitive, and non-radioactive colorimetric method, was used to determine the number of living cells in cell proliferation and cytotoxicity tests (31,32). The A549 human lung epithelial cell line was resuscitated in a dulbecco’s modification of Eagle’s medium (DMEM) medium containing 10% FBS. After two passages, the cells were mixed with trypan blue dye 1:1, the counter was set to “small cell”, and a dilution ratio of 1:2 was applied to count the cells. Cells were diluted to 1×105/mL and seeded into 96-well tissue culture plates with 100 µL per well. The blank group (only medium) and control group (only A549 in medium without bacteria) were set up (4–5 replicates in each group). The cells were cultured at 37 ℃ for 24 h for complete adhesion to the wall. Bacterial infection was carried out according to a MOI =1 (for K. pneumonia) or MOI =25 (for M. smegmatis), and cell viability was measured at 2, 18, 24, and 48 h post-infection according to the instructions recommended by the kit. Briefly, 10 µL of CCK8 solution was added to 100 µL of fresh DMEM medium per well. After incubation at 37 ℃ for 40 min in dark, the absorbance at 450 nm was detected.
Cell viability (%) = [a (test group) − A (blank)]/[a (control group) − (blank)] × 100%.
Western blot
By RIPA lysis buffer (Sigma-Aldrich, USA), total protein in cells was extracted. Following that, proteins were separated using SDS-PAGE, and protein transfer was accomplished using PVDF membranes. In 5% non-fat milk powder, membranes were occluded for 1 hour. After that, membranes were incubated with primary antibodies anti-cleaved caspase-3 (1:500, ab32042, Abcam, USA) and anti-β-actin (1:1,000, ab8227, Abcam) at 4 ℃ overnight. For a further hour at 25 ℃, membranes were cultured with anti-rabbit IgG (ab150077, Abcam). For developing, ECL Ultra Western HRP Substrate (Sigma-Aldrich, USA) was employed. Gray level analysis was done using Image J (NIH, USA).
Immunofluorescence microscopic analysis
Immunofluorescence staining of the K. pneumoniae or M. smegmatis-infected A549 cells was performed to detect the cytoskeleton and cell morphology and then evaluate the cytotoxicity of the two pathogens. Briefly, on sterile 20 mm glass coverslips (Corning, New York), the A549 cells were planted and given 24 hours to reach 80% confluency. After that, the cells were exposed to either M. smegmatis at a MOI of 25:1 for 24 hours or K. pneumonia at a MOI of 1:1 for 24 hours. The media was then removed and fresh RPMI 1640 medium was added. The infected cells were then fixed for 30 minutes with 4% fresh paraformaldehyde, followed by 30 minutes of blocking with 0.5% (w/v) bovine serum albumin (BSA) in PBS. The fixed infected cells were stained by 100 µL Phalloidin-iFluor 647 work solution (red, ab176759; abcam, UK; dilution 1:800) and Actin-Tracker-Green-488 (green, C2201S; Beyotime, China; dilution 1:200) for 60 min at 37 ℃ for actin cytoskeleton staining and 100 µL 4’,6-diamidino-2-phenylindole (DAPI, Life Technologies, Germany) for 15 minutes at 37 ℃ to stain the nucleus. After washing with PBS three times, the samples were then mounted by adding 100 µL of anti-fluorescence quenching agent and observed using a fluorescence microscope (Olympus, Hamburg, Germany).
Transcriptome analysis of the A549 human lung epithelial cell line
The A549 cells were infected with K. pneumonia at a MOI of 1 or M. smegmatis at a MOI of 25 for 24 h. The cell pellets were then suspended in 1 mL of TRIzol reagent (CW Bio, China). RNA extraction was performed as previously described (33,34). Three replicates of each group were performed and a total of 12 samples were prepared for sequencing, including two infection groups and two control groups for different bacteria. Genomic DNA was removed using a PrimerScript™ RT reagent kit (Takara, Japan), and the RNA was precipitated with ethanol and quantified using a Nanodrop (Thermo Fisher Scientific, USA). A total of 2.5 µg RNA per sample was processed into complementary DNA (cDNA) libraries using an Illumina TruSeq RNA sample preparation kit (Illumina, USA) according to the manufacturer’s instructions (Illumina part no. 15026495 Rev. D), and 2×150 bp sequencing was performed on an Illumina Novaseq 6000 instrument. Sequencing was performed using an Illumina Novaseq 6000 platform at Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China).
The raw sequence data was filtered to remove the low-quality reads. The filtered data were aligned against the human reference genome (http://asia.ensembl.org/Homo_sapiens/Info/Index) using TopHat (V2.0.13) (34) with default parameters. The gene expression levels were calculated by the fragments per kilobase of exon per million mapped reads (FRKM) method. The differential expression (DE) of transcripts was established using Cuffdiff (http://cole-trapnell-lab.github.io/cufflinks/cuffdiff/index.html). The Venn diagram was constructed to achieve an overview of the DE gene transcripts present in one or more of the experimental groups. Gene ontology enrichment was analyzed by GOatools. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment of the DE genes was done by KOBAS (http://kobas.cbi.pku.edu.cn/home.do).
Cytokine and chemokine profile determination
Cell culture medium was collected by centrifugation after A549 cells were infected with K. pneumonia at a MOI of 1:1 or M. smegmatis at a MOI of 25:1. Three replicates of each group were performed and a total of nine samples were prepared for determination. The control bacteria included PBS-treated cells (as a negative control). The Bio-Plex® 200 System and the Bio-PlexTM Human Cytokine Standard 27-Plex, Group I (Bio-Rad, Hercules, California, USA) were used to detect cytokine levels in the supernatant at 3 and 24 h post-infection.
Statistical analysis
Each experiment was carried out three times. The means and standard error of mean (SEM) were used to express the results. The analysis of variance (ANOVA) test was applied to analyze statistical significance, followed by Bonferroni multiple comparison post-test. Pearson’s linear correlation coefficient was used to assess the interdependence of variables. P<0.05 was considered statistically significant.
Results
Morphology and growth characteristics of K. pneumoniae and M. smegmatis in vitro
As an Enterobacteriaceae and fast-growing NTM, K. pneumoniae and M. smegmatis showed intrinsic fast proliferation in the 37 ℃ shaker (Figure 1). The growth curve reflected the typical S-type growth characteristics in vitro. Comparatively, the growth rate of K. pneumonia was more rapid. When the initial OD was 0.05, K. pneumonia only needed 3 hours to enter the early logarithmic period, while M. smegmatis needed 12 hours. Although the time to reach a stable period (OD of about 2.5) was close, it is obvious that M. smegmatis can maintain a stable period for a longer time (more than 30 h), while K. pneumonia can maintain this period for less than 10 h. This difference is closely related to the structure of the two bacteria. As a Gram-positive bacterium, Mycobacterium has a lipid-rich hydrophobic cell wall structure, which is important for biofilm formation and harboring drug-tolerant cells (35-37). Gram-negative bacteria do not have a thickened cell wall; however, K. pneumoniae expresses two critical antigens on its cell surface, which contribute to higher levels of adherence and invasion of lung cells and are related to pathogenicity and antibiotic resistance. The precise role of cell wall components in adhesion and invasion remains to be defined (37), but non-pathogenic Escherichia coli and opportunistic pathogenic K. pneumoniae, which cause urinary tract infections, have been shown to form biofilm in mice or in vitro (38,39). Scanning electron microscopy observations have confirmed the physiological base of the cell wall surface. K. pneumoniae forms a regular and intact morphology (Figure 1A), which is uniform in size and shape and connected to the adjacent cells. There are mucinous junctions between bacterial cells at 50,000× magnification. When grown in liquid media, the M. smegmatis strain formed serpentine cords (Figure 1B), which are characteristic of pathogenic mycobacteria such as MTB (40,41). These results further showed that the cell wall integrity and cell morphology were characteristic (42).

Invasion and intracellular survival assays of K. pneumoniae and M. smegmatis
The principal site of entry for pathogenic bacteria into the lungs is the lung epithelium. Adhesion and invasion of the lung epithelial cells is an early stage of the pneumonia process in many respiratory pathogens, including K. pneumoniae. We compared the ability to invade A549 lung epithelial cells and then evaluated the cytotoxicity of K. pneumoniae and M. smegmatis in this section, and the results are shown in Figure 2.
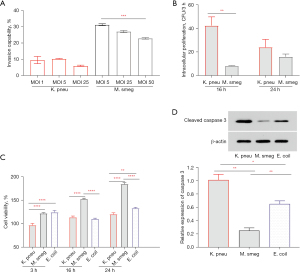
A549 lung epithelial cells were infected for 3 h with different MOIs. The efficiency of invasion referred to the rate of entering cells, which was calculated by dividing the number of intracellular bacteria by the number of initially infected bacteria. We compared three MOIs (MOI =1, 5, and 25 for K. pneumonia and MOI =5, 25, and 50 for M. smegmatis) to investigate whether the invasion was dependent on the number of bacteria (Figure 2A). In K. pneumonia infection, the rate of entering cells declined from 9.52% (MOI =1) to 6.03% (MOI =25). In M. smegmatis infection, the rate of entering cells declined from 31.25% (MOI =5) to 22.89% (MOI =50). It was suggested that excessive infection may prevent bacteria from entering lung epithelial cells or even damaging cells. In general, the ability to invade epithelial cells is higher in M. smegmatis; this is consistent with the morphological characteristics of serpentine cords, which increase bacterial adhesion to host cells. For intracellular pathogens, the number of viable bacteria will directly affect the cellular immune response and subsequently determine the outcome of infection. Viable bacteria in the cells were determined to be 5.32Log10 (CFU) K. pneumonia at a MOI =1 and 5.88Log10 (CFU) M. smegmatis at a MOI =25, so we selected these MOIs of two bacteria for the subsequent experiments in this study. Viable intracellular bacteria continued to increase over time by enumerating the bacterial number (colony forming unit, CFU) at the 16 and 24 h timepoints post-infection, as shown in Figure 2B. Relatively, M. smegmatis grew more slowly during the early stage in A549 cells. However, it seemed that the high growth ability of K. pneumonia cannot be sustained because intracellular CFU of the two bacteria tended to be the same at 24 h post-infection.
To determine whether K. pneumoniae and M. smegmatis affected the proliferation and activity of host cells, CCK-8 was used to detect such changes along with the infection time. The results showed that the activity of CCK-8 in the M. smegmatis-infected group continued to increase from 3 to 24 h post-infection, but there was no significant difference at each time point in the K. pneumoniae-infected group (Figure 2C). Compared to the two infected groups at the same time point, the cell viability of the M. smegmatis-infected group was significantly higher than that of the K. pneumoniae-infected group. In particular, Escherichia coli, a non-pathogenic strain of Enterobacteriaceae with cell adhesion, was selected as the control infection group of K. pneumoniae. There are no magnificent differences between the A549 cytotoxicities between K. pneumonia and Escherichia coli. Furthermore, western blot was performed to detected activity of apoptosis related protein cleaved caspase-3. The result showed that the level of cleaved caspase-3 in M. smegmatis-infected group was significantly lower than that in K. pneumoniae-infected group and E. coli-infected group at 24 h post-infection. Taken together, these findings indicated that K. pneumoniae and M. smegmatis infection did not affect the viability and proliferation of epithelial cells, and the presence of M. smegmatis seemed to be beneficial to cell proliferation over time.
Immunofluorescence test for F-actin
The cell cytoskeleton can be manipulated by microbial pathogens to facilitate productive infection (43-45). For example, K. pneumoniae can induce a cytotoxic effect in lung epithelial cells with a high MOI ranging from 100:1 to 1,000:1, and also increases Factin protein levels in mouse lung tissue (18,25). Mycobacterial adhere to the host epithelium with MBP-1 protein by interacting with the host cytoskeletal protein vimentin (9). We then compared the F-actin cytoskeleton protein levels at lower MOIs (Figure 3). F-actin staining revealed that the cell morphology and cytoskeleton could not be changed by low-dose K. pneumoniae (Figure 3A) and M. smegmatis (Figure 3B) infections. Many infections enter the body through the lung, where they may easily penetrate the alveolar-capillary barrier and enter the circulation. Diverse mechanisms take part in protecting the lung, and the number of normal symbiotic microorganisms will not be extremely high, except in cases of acute infection. Therefore, we chose the low dose of infection to perform this research, and the results were more conducive to imitating the natural infection state.

Transcription pattern of lung epithelial cells after K. pneumoniae and M. smegmatis infection
To explore the transcriptional regulation mechanism of A549 lung epithelial cells infected by K. pneumoniae and M. smegmatis, we chose the 24 h time point to develop RNA-sequencing (RNA-seq) based on viable bacteria-cell changes after infection. A total of 12 samples were collected, comprising four groups: two bacterial-infection groups and two respective controls. A high ratio (>96%) and sequencing depth indicated that the RNA-seq data was high quality. We successfully identified more than 10,000 genes in each sample and obtained the gene expression matrix (Figure 4A). We then analyzed the correlation between the two infection groups to evaluate the host differences based on different pathogens (Figure 4B). As shown in Figure 4, in terms of the biological view, significant differences between the two groups indicated that the expression trend of most genes in A549 cells was different. We identified the differentially expressed genes (DEGs) according to the following criteria: fold change (FC) >2 or <0.5; FDR P<0.05.
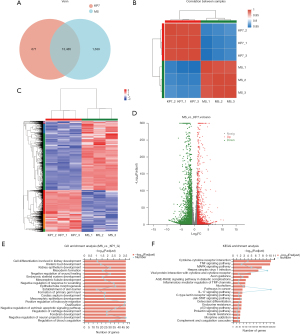
The list of DEGs was shown in website: https://cdn.amegroups.cn/static/public/jtd-23-493-1.xlsx. The analysis of the global gene regulation profiles of A549 cells treated with K. pneumoniae showed that 4,755 genes were significantly differentially expressed, including 1,797 up-regulated and 2,958 down-regulated genes (Figure 4C,4D).
We conducted a gene ontology (GO) enrichment analysis of 4,755 DEGs based on the GO database in order to evaluate the functional implications of the changes in gene expression brought on by K. pneumoniae exposure. Figure 4E listed the top 20 ranked GO terms of the DEGs. The main biological processes were cell differentiation involved in kidney development, ureteric bud development, kidney epithelium development, mesoderm formation, negative regulation of wound healing, embryonic skeletal system development, and mesonephric tubule development, suggesting that the processes of A549 cells were most influenced function with K. pneumoniae infection.
Seventy-eight pathways with DEs (P<0.05) were accessed between the treatment and control groups, among which 22 pathways exhibited the most notable differences (P<0.01), and the top 20 ranked significant pathways in KEGG were exhibited (Figure 4F). Numerous signal transduction pathways were enriched, containing the tumor necrosis factor (TNF) pathway, the MAPK pathway, viral protein interaction with cytokines and cytokine receptors, the IL-17 signaling pathway, the Janus kinase-signal transducer and activator of transcription (Jak-STAT) signaling pathway, and the C-type lectin receptor signaling pathway. In addition, biological processes regulation such as cytokine-cytokine receptor interaction, herpes simplex virus 1 infection, endocrine resistance, and insulin resistance were identified in our results. The results provided vital information for the exploration of K. pneumoniae in A549 cells.
Cytokine and chemokine profile determination
Recombinant pure outer membrane protein A (OmpA) from K. pneumoniae has been demonstrated in several investigations to trigger the production of inflammatory molecules in a TLR2-dependent way in a variety of cell types (46). The TLR signaling pathway is vital in resisting pathogenic bacterial invasions (such as MTB), stimulating the production of cytokines, and activating the immune response (47). Epithelial cells release TNF-α, interferon-γ (IFN-γ), chemokines, and other cytokines, which respond to Mycobacterium infection. A study has found that M. smegmatis can be transformed into a pathogenic phenotype via A549 epithelial cells during macrophage infection (48). To compare the DE of immune factors, the cytokine patterns induced were compared to find the specific cytokines related to K. pneumonia (MOI =1) and M. smegmatis (MOI =25). Statistical analysis revealed significant changes in the levels of various cytokines (Figure 5A). IL-8 and monocyte chemoattractant protein-1 (MCP-1) were secreted at high levels at both 3 and 24 h after infection with K. pneumonia and M. smegmatis (Figure 5B,5C). The significantly increased expression of IL-8 and MCP-1 was observed in the M. smegmatis infection group at both time points among the three groups. Meanwhile, K. pneumonia infection induces a significant decrease in MCP-1 secretion compared with the control group. Similarly, IL-1α and interferon gamma-induced protein 10 (IP-10) was decreased in the K. pneumonia infection group at 3 h post-infection. The levels of certain cytokines (IL-1α, IL-5, IL-9, IL-10, and IL-15), chemokines [IP-10, fibroblast growth factor (FGF), granulocyte colony-stimulating factor (G-CSF), vascular endothelial growth factor (VEGF)], platelet-derived growth factor bb (PDGFF-bb), all were significantly increased in the two infection groups at 24 h post-infection. The comparison between the two infection groups at 24 h revealed increased secretion of VEGF, FGF, G-CSF, and IL-1α in the K. pneumonia infection group; meanwhile, in the M. smegmatis infection group, IL-5 and IL-6 were regulated on activation, normal T cells expressed and secreted (RANTES) chemokine, and there was increased secretion of PDGF-bb (Figure 5D). The Bio-Plex® assay revealed statistically significant differences between the infection and control groups in the levels of the above cytokines, chemokines, and growth factors (Figure 5). Another 13 cytokines were not shown, either because there was no significant difference among the three groups or the expression level was too low to be detected.

Discussion
Although the immunopathological mechanisms of K. pneumoniae and M. smegmatis in lung disease have not been clarified, our results suggest that epithelial cells should not be thought of as just a physical barrier against infection. In this respect, we have shown the following. Firstly, in lung epithelial cells, pathogens are allowed to stop and proliferate. Epithelial cells regulate the secretion of immune factors at the gene transcription level and produce different immune responses depending on the invasion of different pathogens. Secondly, the persistence and systemic spread of these opportunistic infections are caused by the capacity of K. pneumoniae and M. smegmatis strains to build biofilm and infiltrate epithelial cells. In contrast, M. smegmatis has a stronger adhesion and intracellular retention ability, while K. pneumoniae is more likely to induce acute infection.
Recent research from several labs has emphasized the significance of bacterial interactions across species in regulating bacterial pathogenicity and the responsiveness to antibiotic treatment in the pulmonary infections of both CF and non-CF patients (49,50). In CF patients, pulmonary tissues are usually considered always polymicrobial (51). This study, represented by K. pneumoniae and M. smegmatis, compared the infection mechanisms of Gram-positive and Gram-negative bacteria in lung epithelium cells, provided basic research data for the control of mixed bacterial infection in the lung, and also provided a new perspective for novel intervention measures.
Both K. pneumoniae and M. smegmatis were reported to have an in vivo biofilm formation ability, including increased antibiotic resistance and a significant ability to evade host defense factors. Our results showed that the cell cluster morphology (M. smegmatis) and intercellular adhesion (K. pneumoniae) were strongly suggestive of biofilm formation. Unlike macrophages, pulmonary epithelial cells have no active phagocytosis; such morphology will help pathogen entry into A549 cells. The following adhesion and endocytosis experiments confirmed our hypothesis. Significantly, M. smegmatis invades epithelial cells more efficiently, indicating that compared with the lipopolysaccharide (LPS) and capsular polysaccharides of Gram-negative bacteria, the unique cord factor structure of the cell wall of Mycobacterium is more conducive to the invasion and even spread of bacteria. Although both K. pneumoniae and M. smegmatis both exhibited an ability to grow in epithelial cells, the difference in intracellular proliferation ability between them is very significant, which is consistent with their growth curve in vitro. M. smegmatis showed a stronger long-term survival ability, which was consistent with its longer stability phase in vitro. These results suggested that M. smegmatis can persist in epithelial cells for a long time, known as latent infection. At present, one-fourth of the world’s population has a latent MTB infection, causing huge hidden dangers from tuberculosis and the spread of drug-resistant bacteria (52). Even more worrying is that once these bacteria leave the epithelial cells and infect other cells, their virulence will be greatly enhanced (48).
Numerous cellular activities, such as endocytosis, motility, nutrition uptake, and mitosis, depend on the cytoskeleton, a highly dynamic structural framework. By manipulating the cytoskeleton, intracellular and extracellular pathogenic bacteria can induce host cell-take up, steal resources from host organelles, and eventually create a niche for their own reproduction. In the present study, no change in F-actin was observed within 24 hours after infection by immunofluorescence. However, the RNA-seq results showed that the expression of cytoskeleton-related proteins differed between the two infected cells, including the increased transcription levels of cytoskeletal calmodulin and titin-interacting Rho family guanine nucleotide exchange factor (RhoGEF), fascin actin-bundling protein 3 (FSCN3), actin-related protein T3, actin-binding LIM protein family member 2 (ABLIM2), and so on in pulmonary epithelial cells infected with K. pneumonia. PI3K is the central enzyme of many signaling cascades and can promote the recruitment of effectors, which further activate receptors and link with actin cytoskeleton rearrangement. We did not observe an expression change in the RNA data; however, another transcription regulator (TF), AKT, the main effector downstream of PI3K, was up-regulated in the K. pneumonia infection group, which partly explained the genetic basis of the actin changes. In general, the changes in cytoskeleton-related proteins are worthy of further detection to facilitate investigation regarding the pathological significance of these changes after K. pneumoniae and M. smegmatis infection, especially in epithelial cells which remains largely unexplored.
In response to pathogen invasion, cytokines and chemokines are essential chemicals that play a role in practically every aspect of immunity and inflammation. The identification of pathogens by airway epithelial cells, which activates signaling pathways and causes the production of co-stimulatory molecules as well as the release of cytokines and chemokines, is a critical step in the lung’s defense against infections. Evidence suggests that K. pneumoniae infections are distinguished by the lack of an early inflammatory response. Nevertheless, it remains uncertain whether K. pneumoniae employs other agents to control inflammatory reactions in the host. IL-6 has been found to improve the survival of K. pneumoniae-induced pneumonia sepsis patients by boosting neutrophil death. It has also been shown that CXCL1 can increase resistance to K. pneumonia in mice. Moreover, lipocalin 2 (LCN2) may restrict bacterial growth by securing the iron-binding bacterial siderophores and preventing bacterial access to iron. In addition to its role in the dissemination of M. tuberculosis, epithelial cells release TNF-α, IFN-γ, chemokines, and other cytokines in response to mycobacterium infection (53,54). A recent study revealed that K. pneumonia might induced acute lung injury by increased the level of IL-6, CXCL1, CXCL2, and caspase-9 (55). In addition, M. smegmatis activates host inflammation by increasing expression of TNF-α, IL-1β, and IL-6, while suppressing the expression of anti-inflammatory cytokines through the NFκB, ERK1/2, and p38 mitogen-activated protein kinase pathways, which causes acute lung injury (56). It is likely that the cytokines essential for optimal bacterial clearance will also differ. Since bacterial loads may likely cause severe tissue harm and death over time, we examined the cytokine levels early in the current investigation. Most cytokines could not be detected due to their low concentrations, except for MCP-1, IL-8, IL-1Ra, IP-10, and VEGF at 3 h post-infection. Among them, VEGF expression was significantly higher in both K. pneumonia and M. smegmatis infection groups than that of the control at 3 and 24 h. We also found that low-dose pathogenic microorganism infection may promote pulmonary epithelial cells to release VEGF and then could change the composition of local immune cells in the lung. The secretion levels of pro-inflammatory cytokines, such as MCP-1, IL-8, and IL-1Ra, together with PDGFF-bb were significantly more induced by M. smegmatis than by K. pneumonia. With its ability to activate neutrophils’ respiratory burst and defensin release via CXCR1, IL-8 is crucial in controlling the acute inflammatory response to invading pathogens. The induction effect of M. smegmatis on MCP-1, IL-6, and IL-8 increased time-dependently, implying that M. smegmatis infection can promote crosstalk from a very early stage among epithelial cells and other immune cells, including monocytes, macrophages, etc. K. pneumonia can induce the secretion of immune factors from epithelial cells relatively slowly. A549 cells may be more inclined to respond to early K. pneumonia infection by altering the cytoskeletal proteins. Significant correlations between K. pneumonia infection and higher levels of IL-15, IL-1Ra, FGF basic, and G-CSF secretion were also observed.
Conclusions
Research has shown that exposure to cigarette extract suppresses the release of many pro-inflammatory cytokines, including IL-6, IL-8, IP-10, and RANTES, while other studies have found that it induces the production of IL-1β and IL-8 in human bronchial epithelial cells (HBE-14o), which highlights the necessity and advantage of the head-to-head comparison conducted in this study. To verify our findings, more research with in vivo models that more closely resemble pulmonary tissues is required. The roles of these two bacteria in the activation of the genes required for intracellular survival and presentation their pathogenicity needs further study. Furthermore, co-infection with a 2–3 pathogens model based on the findings in this study may help develop new strategies for the prevention and treatment of pulmonary infection.
Acknowledgments
Funding: This research was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-493/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-493/dss
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-493/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-493/coif). The authors have no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Holden VI, Breen P, Houle S, et al. Klebsiella pneumoniae Siderophores Induce Inflammation, Bacterial Dissemination, and HIF-1alpha Stabilization during Pneumonia. mBio 2016;7:e01397-16. [Crossref] [PubMed]
- Dell'Annunziata F, Ilisso CP, Dell'Aversana C, et al. Outer Membrane Vesicles Derived from Klebsiella pneumoniae Influence the miRNA Expression Profile in Human Bronchial Epithelial BEAS-2B Cells. Microorganisms 2020;8:1985. [Crossref] [PubMed]
- Buonocore C, Tedesco P, Vitale GA, et al. Characterization of a New Mixture of Mono-Rhamnolipids Produced by Pseudomonas gessardii Isolated from Edmonson Point (Antarctica). Mar Drugs 2020;18:269. [Crossref] [PubMed]
- Lee CR, Lee JH, Park KS, et al. Antimicrobial Resistance of Hypervirulent Klebsiella pneumoniae: Epidemiology, Hypervirulence-Associated Determinants, and Resistance Mechanisms. Front Cell Infect Microbiol 2017;7:483. [Crossref] [PubMed]
- Paczosa MK, Mecsas J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol Mol Biol Rev 2016;80:629-61. [Crossref] [PubMed]
- Lin JE, Valentino M, Marszalowicz G, et al. Bacterial heat-stable enterotoxins: translation of pathogenic peptides into novel targeted diagnostics and therapeutics. Toxins (Basel) 2010;2:2028-54. [Crossref] [PubMed]
- Wang H, Zhong Z, Luo Y, et al. Heat-Stable Enterotoxins of Enterotoxigenic Escherichia coli and Their Impact on Host Immunity. Toxins (Basel) 2019;11:24. [Crossref] [PubMed]
- Ren M, Li L, Chu M, et al. Detection of Klebsiella pneumoniae cfDNA in pleural fluid and its clinical value. Ann Palliat Med 2020;9:3379-84. [Crossref] [PubMed]
- Babrak L, Danelishvili L, Rose SJ, et al. The environment of "Mycobacterium avium subsp. hominissuis" microaggregates induces synthesis of small proteins associated with efficient infection of respiratory epithelial cells. Infect Immun 2015;83:625-36. [Crossref] [PubMed]
- Miltner E, Daroogheh K, Mehta PK, et al. Identification of Mycobacterium avium genes that affect invasion of the intestinal epithelium. Infect Immun 2005;73:4214-21. [Crossref] [PubMed]
- Martini MC, Zhou Y, Sun H, et al. Defining the Transcriptional and Post-transcriptional Landscapes of Mycobacterium smegmatis in Aerobic Growth and Hypoxia. Front Microbiol 2019;10:591. [Crossref] [PubMed]
- Alqurashi MM, Alsaileek A, Aljizeeri A, et al. Mycobacterium smegmatis causing a granulomatous cardiomediastinal mass. IDCases 2019;18:e00608. [Crossref] [PubMed]
- Al-Ghafli H, Al-Hajoj S. Nontuberculous Mycobacteria in Saudi Arabia and Gulf Countries: A Review. Can Respir J 2017;2017:5035932. [Crossref] [PubMed]
- Brown-Elliott BA, Philley JV. Rapidly Growing Mycobacteria. Microbiol Spectr 2017;
- Shimizu F, Hatano Y, Okamoto O, et al. Mycobacterium smegmatis soft tissue infection. Int J Dermatol 2012;51:1518-20. [Crossref] [PubMed]
- Saffo Z, Ognjan A. Mycobacterium smegmatis infection of a prosthetic total knee arthroplasty. IDCases 2016;5:80-2. [Crossref] [PubMed]
- Namouchi A, Cimino M, Favre-Rochex S, et al. Phenotypic and genomic comparison of Mycobacterium aurum and surrogate model species to Mycobacterium tuberculosis: implications for drug discovery. BMC Genomics 2017;18:530. [Crossref] [PubMed]
- Zheng S, Ren L, Li H, et al. High-mobility group nucleosome-binding domain 2 protein inhibits the invasion of Klebsiella pneumoniae into mouse lungs in vivo. Mol Med Rep 2015;12:1279-85. [Crossref] [PubMed]
- Borish LC, Steinke JW. 2. Cytokines and chemokines. J Allergy Clin Immunol 2003;111:S460-75. [Crossref] [PubMed]
- Sutherland RE, Olsen JS, McKinstry A, et al. Mast cell IL-6 improves survival from Klebsiella pneumonia and sepsis by enhancing neutrophil killing. J Immunol 2008;181:5598-605. [Crossref] [PubMed]
- Cai S, Batra S, Lira SA, et al. CXCL1 regulates pulmonary host defense to Klebsiella Infection via CXCL2, CXCL5, NF-kappaB, and MAPKs. J Immunol 2010;185:6214-25. [Crossref] [PubMed]
- Ehlers S. Lazy, dynamic or minimally recrudescent? On the elusive nature and location of the mycobacterium responsible for latent tuberculosis. Infection 2009;37:87-95. [Crossref] [PubMed]
- Lemos MP, McKinney J, Rhee KY. Dispensability of surfactant proteins A and D in immune control of Mycobacterium tuberculosis infection following aerosol challenge of mice. Infect Immun 2011;79:1077-85. [Crossref] [PubMed]
- Gaglione R, Cesaro A, Dell'Olmo E, et al. Cryptides Identified in Human Apolipoprotein B as New Weapons to Fight Antibiotic Resistance in Cystic Fibrosis Disease. Int J Mol Sci 2020;21:2049. [Crossref] [PubMed]
- Cano V, Moranta D, Llobet-Brossa E, et al. Klebsiella pneumoniae triggers a cytotoxic effect on airway epithelial cells. BMC Microbiol 2009;9:156. [Crossref] [PubMed]
- Faron M, Fletcher JR, Rasmussen JA, et al. Interactions of Francisella tularensis with Alveolar Type II Epithelial Cells and the Murine Respiratory Epithelium. PLoS One 2015;10:e0127458. [Crossref] [PubMed]
- Wu Q, Jiang D, Minor MN, et al. In vivo function of airway epithelial TLR2 in host defense against bacterial infection. Am J Physiol Lung Cell Mol Physiol 2011;300:L579-86. [Crossref] [PubMed]
- McCormick TS, Weinberg A. Epithelial cell-derived antimicrobial peptides are multifunctional agents that bridge innate and adaptive immunity. Periodontol 2000 2010;54:195-206. [Crossref] [PubMed]
- Kato A, Schleimer RP. Beyond inflammation: airway epithelial cells are at the interface of innate and adaptive immunity. Curr Opin Immunol 2007;19:711-20. [Crossref] [PubMed]
- Vadivelu J, Vellasamy KM, Thimma J, et al. Survival and Intra-Nuclear Trafficking of Burkholderia pseudomallei: Strategies of Evasion from Immune Surveillance? PLoS Negl Trop Dis 2017;11:e0005241. [Crossref] [PubMed]
- Lu Z, Zou J, Li S, et al. Epigenetic therapy inhibits metastases by disrupting premetastatic niches. Nature 2020;579:284-90. [Crossref] [PubMed]
- Luther A, Urfer M, Zahn M, et al. Chimeric peptidomimetic antibiotics against Gram-negative bacteria. Nature 2019;576:452-8. [Crossref] [PubMed]
- Saini V, Farhana A, Steyn AJ. Mycobacterium tuberculosis WhiB3: a novel iron-sulfur cluster protein that regulates redox homeostasis and virulence. Antioxid Redox Signal 2012;16:687-97. [Crossref] [PubMed]
- Jiang J, Lin C, Zhang J, et al. Transcriptome Changes of Mycobacterium marinum in the Process of Resuscitation From Hypoxia-Induced Dormancy. Front Genet 2019;10:1359. [Crossref] [PubMed]
- Hett EC, Rubin EJ. Bacterial growth and cell division: a mycobacterial perspective. Microbiol Mol Biol Rev 2008;72:126-56. table of contents. [Crossref] [PubMed]
- Vijay S, Nagaraja M, Sebastian J, et al. Asymmetric cell division in Mycobacterium tuberculosis and its unique features. Arch Microbiol 2014;196:157-68. [Crossref] [PubMed]
- Li X, He J, Fu W, et al. Effect of Mycobacterium tuberculosis Rv3717 on cell division and cell adhesion. Microb Pathog 2018;117:184-90. [Crossref] [PubMed]
- Van Laar TA, Chen T, You T, et al. Sublethal concentrations of carbapenems alter cell morphology and genomic expression of Klebsiella pneumoniae biofilms. Antimicrob Agents Chemother 2015;59:1707-17. [Crossref] [PubMed]
- Tsuneda S, Aikawa H, Hayashi H, et al. Extracellular polymeric substances responsible for bacterial adhesion onto solid surface. FEMS Microbiol Lett 2003;223:287-92. [Crossref] [PubMed]
- Wu J, Ru HW, Xiang ZH, et al. WhiB4 Regulates the PE/PPE Gene Family and is Essential for Virulence of Mycobacterium marinum. Sci Rep 2017;7:3007. [Crossref] [PubMed]
- Ren H, Dover LG, Islam ST, et al. Identification of the lipooligosaccharide biosynthetic gene cluster from Mycobacterium marinum. Mol Microbiol 2007;63:1345-59. [Crossref] [PubMed]
- He J, Fu W, Zhao S, et al. Lack of MSMEG_6281, a peptidoglycan amidase, affects cell wall integrity and virulence of Mycobacterium smegmatis. Microb Pathog 2019;128:405-13. [Crossref] [PubMed]
- Carabeo RA, Grieshaber SS, Fischer E, et al. Chlamydia trachomatis induces remodeling of the actin cytoskeleton during attachment and entry into HeLa cells. Infect Immun 2002;70:3793-803. [Crossref] [PubMed]
- Elliott DA, Clark DP. Cryptosporidium parvum induces host cell actin accumulation at the host-parasite interface. Infect Immun 2000;68:2315-22. [Crossref] [PubMed]
- Elliott DA, Coleman DJ, Lane MA, et al. Cryptosporidium parvum infection requires host cell actin polymerization. Infect Immun 2001;69:5940-2. [Crossref] [PubMed]
- March C, Moranta D, Regueiro V, et al. Klebsiella pneumoniae outer membrane protein A is required to prevent the activation of airway epithelial cells. J Biol Chem 2011;286:9956-67. [Crossref] [PubMed]
- Sequeira PC, Senaratne RH, Riley LW. Inhibition of toll-like receptor 2 (TLR-2)-mediated response in human alveolar epithelial cells by mycolic acids and Mycobacterium tuberculosis mce1 operon mutant. Pathog Dis 2014;70:132-40. [Crossref] [PubMed]
- Kim SY, Sohn H, Choi GE, et al. Conversion of Mycobacterium smegmatis to a pathogenic phenotype via passage of epithelial cells during macrophage infection. Med Microbiol Immunol 2011;200:177-91. [Crossref] [PubMed]
- Pompilio A, Crocetta V, Confalone P, et al. Adhesion to and biofilm formation on IB3-1 bronchial cells by Stenotrophomonas maltophilia isolates from cystic fibrosis patients. BMC Microbiol 2010;10:102. [Crossref] [PubMed]
- Hoffman LR, Déziel E, D'Argenio DA, et al. Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 2006;103:19890-5. [Crossref] [PubMed]
- Sibley CD, Rabin H, Surette MG. Cystic fibrosis: a polymicrobial infectious disease. Future Microbiol 2006;1:53-61. [Crossref] [PubMed]
- World Health Organization. Global tuberculosis report 2020. Geneva: World Health Organization; 2020.
- Awuh JA, Flo TH. Molecular basis of mycobacterial survival in macrophages. Cell Mol Life Sci 2017;74:1625-48. [Crossref] [PubMed]
- Sia JK, Rengarajan J. Immunology of Mycobacterium tuberculosis Infections. Microbiol Spectr 2019;7: [Crossref] [PubMed]
- Jiang W, Liu J, Zhao X, et al. Melatonin ameliorates lung cell inflammation and apoptosis caused by Klebsiella pneumoniae via AMP-activated protein kinase. Inflammopharmacology 2022;30:2345-57. [Crossref] [PubMed]
- Suo J, Wang X, Zhao R, et al. Mycobacterium tuberculosis PPE7 Enhances Intracellular Survival of Mycobacterium smegmatis and Manipulates Host Cell Cytokine Secretion Through Nuclear Factor Kappa B and Mitogen-Activated Protein Kinase Signaling. J Interferon Cytokine Res 2022;42:525-35. [Crossref] [PubMed]


