
4-phenylbutyric acid re-trafficking hERG/G572R channel protein by modulating the endoplasmic reticulum stress-associated chaperones and endoplasmic reticulum-associated degradation gene
Highlight box
Key findings
• 4-PBA is expected to be developed as a new treatment method for LQT2/G572R. 4-PBA corrects hERG channel transport defects by inhibiting excessive endoplasmic reticulum stress (ERS) and the endoplasmic reticulum (ER)-associated degradation (ERAD)-related gene E3 ubiquitin ligase HRD1. Additionally, 4-PBA improved WT/G572R channel current.
What is known and what is new?
• hERG/G572R channel has been known to induce ERS.
• A novel finding is that 4-PBA may be therapeutic by inhibiting the persistent excessive ERS response and ERAD gene HRD1 of the hERG/G572R channel.
What is the implication, and what should change now?
• This suggests that 4-PBA may become a new drug for treating LQT2/G572R, but more large-scale multi-center clinical trials are needed to verify it.
Introduction
Long QT syndrome (LQTS) is a kind of cardiac ion channel disease caused by genetic mutation. It is characterized by arrhythmia, syncope, and sudden cardiac death (SCD). Congenital long QT syndrome (cLQTS) is characterized by prolonged QT interval and prolonged action potential duration (APD). The three major genotypes of LQTS, namely, LQT1, LQT2, and LQT3, account for about 80–90% of LQTS patients. Hundreds of LQT2-related mutations have been described, and the hERG (also called KCNH2) channel encodes the K+ channel alpha subunit that conducts the cardiac rapid delayed rectifier K+ current (IKr). There are four types of diseases causing ion channel mutations: (I) abnormal synthesis; (II) protein transport defects; (III) abnormal ion permeability, and (IV) abnormal gating.
A study has shown that 90% of hERG mutations inhibit kv11.1 transport from the endoplasmic reticulum (ER) to Golgi and cell surface membrane by destroying subunit folding and assembly (1). hERG mutations in LQT2 cause hERG transport disorders by stimulating the endoplasmic reticulum stress (ERS) response, such as point mutation hERG/G572R. This mutation results in the substitution of glycine residue at 572 positions in the transmembrane region of hERG potassium channel S5 with arginine residue. hERG/G572R is located in the transmembrane pore domain (PD) (S5-loop-S6); the incidence of cardiac events in patients with hERG transmembrane pore (S5-loop-S6) missense mutations is significantly higher than that in patients with N-terminal and C-terminal missense mutations. PD mutations in the hERG channel often cause significant physiological consequences after ERS. Among alleles, mutated genes causing less than 50% functional defects are haploinsufficient, and those causing greater than 50% functional defects are dominant negative phenotypes (2). hERG/G572R is a Mendelian inherited disease, and when mutated genes are coexpressed with normally functioning wild type (WT) gene subunits to form alleles, they cause a significant dominant negative suppressor effect. When hERG mutations are coexpressed with WT subunits to form heterotetramers, the dramatic improvement [PD > Per-Arnt-Sim domain (PASD) > C-linker/cyclic nucleotide-binding homology domain (CNBHD)] pharmacology (E4031) correction to escape ER quality control systems [such as ER-associated degradation (ERAD)] is now understood to be more common than previously thought. Most of the transport-deficient but correctable mutations are located in the PD between S5 and S6 (3).
The biogenesis of hERG tetramer and hERG/G572R functional defects are described below (Figure 1). First, hERG DNA is transcribed into hERG messenger RNA (mRNA) in the nucleus and exported to the ER after ribosome translation, where it is folded and assembled into 135 kDa hERG tetramer. Then, correctly folded hERG protein is processed into 155 kDa hERG at the Golgi apparatus and exported to the cell membrane via coat protein complex II (COPII) vesicles to form hERG channel proteins with transmembrane helical structural domains S1–S6. Meanwhile, hERG protein misfolding and hERG protein unfolding after gene mutation (G572R) causes an ERS response. The ERS process is involved in three ER transmembrane proteins: inositol-requiring enzyme 1 (IRE1), activating transcription factor 6 (ATF6), and protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK). When the ER is in a non-stress state, ATF6 binds to a glucose-regulated protein (GRP) molecular chaperone (such as GRP78 and GRP94) to form ATF6-GRP78/GRP94 complex to maintain a quiescent, inactive state (4). During ERS, unfolded or misfolded proteins cause GRP to dissociate from ATF6, triggering the unfolded protein response (UPR). UPR induces the upregulation of ER chaperones such as BIP (also known as GRP78 and HSP5A), GRP94 (also known as HSP90B1), and calnexin (CNX), calreticulin (CRT), which promote proper folding of ER unfolded proteins (5,6). Moreover, misfolded, incompletely folded, and improperly assembled mutant proteins are recognized by ER quality control and cause ERS response. They are confined to the ER, unable to export to the Golgi and cell membrane, and degraded through the ERAD pathway. 3-Hydroxy-3-methylglutaryl coenzyme A reductase degradation protein 1 (HRD1) is a key component of the ERAD pathway (6), which is responsible for misfolded protein recognition in ER and reverse translocation for cytosolic proteasome degradation. The HRD1 pathway is one of the main pathways for the degradation of misfolded ER proteins in eukaryotic cells, which is induced by ERS and up-regulated by the overexpression of ERS sensor molecules IRE1 and ATF6 (7,8).
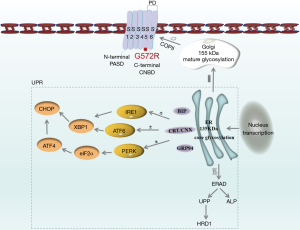
In addition, 4-phenylbutyric acid (4-PBA) low molecular weight fatty acid, which prevents aggregation of misfolded proteins and attenuates ERS-mediated autophagy and apoptosis, has been used to treat excessive ERS (9). 4-PBA has chaperone-like effects in increasing protein stability, contributing to remodeling of unfolded proteins, and has been approved by the USA Food and Drug Administration (FDA) for the prevention and treatment of diseases caused by the accumulation of misfolded proteins (10). Much literature has shown that 4-PBA can reduce the accumulation of ER mutant proteins and restore the physiological function of the mutant protein, which is a promising strategy for allele-specific therapy. For example, 4-PBA rescues some creatine transporter deficiency (CTD) variants of protein misfolding and allows their delivery to the correct subcellular location/trafficking to the plasma membrane (11), and the membrane localization of many dysferlin (DYSF) mutations has been shown to be partially restored by 4-PBA (12). Moreover, 4-PBA is recognized as a typical ERS inhibitor, which assists in protein folding, increases protein stability, and reduces persistent UPR signaling.
In light of the mechanism of ERS caused by hERG mutant proteins and the promising therapeutic effects of the small molecule 4-PBA. In this study, we hypotheses hERG/G572R is capable of being regulated by 4-PBA to re-transport the mutant protein and used experimental analysis to investigate the influence of transport-deficient hERG/G572R mutant protein on ERS-related pathways and ERAD gene. We present this article in accordance with the MDAR reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1252/rc).
Methods
Overview of the experimental procedures
First, the WT, G572R heterozygous and homozygous HEK293 cell lines were constructed. Subsequently, whole-cell patch-clamp experiment were used to verify the channel current of WT/G572R before and after 4-PBA treatment; western blot (WB) experiment were used to verify the protein expression of hERG before and after 4-PBA treatment; and quantitative real-time polymerase chain reaction (qPCR) experiment were used to verify the mRNA expression of hERG before and after 4-PBA treatment; co-immunoprecipitation (co-IP) experiment was used to verify the interaction among hERG, GRP78, GRP94, and HRD1.
Antibodies and drugs
Rabbit polyclonal anti-KCNH2 (hERG) antibody (cat. no. APC-062) was purchased from Alomone Labs (1:200; Jerusalem, Israel). Anti-DDDDK tag (Binds to FLAG® tag sequence) rabbit antibody (EPR20018-251) (1:1,000; cat. no. ab205606) were purchased from Abcam (Cambridge, UK). ATF6 rabbit polyclonal antibody (1:3,000; cat. no. 24169-1-AP), GRP78/BIP rabbit polyclonal antibody (1:3,000; cat. no. 11587-1-AP), GRP94 rabbit polyclonal antibody (1:3,000; cat. no. 14700-1-AP), and HRD1/SYVN1 rabbit polyclonal antibody (1:3,000; cat. no. 13473-1-AP) were purchased from Proteintech® (Wuhan, China). ProteinFind® anti-β-tubulin mouse monoclonal antibody (1:3,000; cat. no. HC101-01) was purchased from TransGen Biotech Co., Ltd. (Beijing, China). ProteinFind® goat anti-rabbit IgG (H+L), horseradish peroxidase (HRP) conjugate (1:3,000; cat. no. HS101-01) was purchased from TransGen Biotech Co., Ltd. (Beijing, China). ProteinFind® goat anti-mouse IgG (H+L), HRP conjugate (1:3,000; cat. no. HS201-01) was purchased from TransGen Biotech Co., Ltd. (Beijing, China). 4-PBA (cat. no. P831259) was purchased from Macklin (Shanghai, China). Insoluble 4-PBA was washed with phosphate-buffered saline (PBS), H2O, and Dulbecco’s Modified Eagle Medium (DMEM). Then, the 4-PBA was heated and/or sonicated. Lipid-soluble 4-PBA was dissolved with dimethyl sulfoxide (DMSO) according to the manufacturer’s instructions. The aim was to prepare the reagents and drugs needed for the experiment.
Plasmid construction, cDNA cloning, and cell culture
Using the pcDNA3 vector to express WT/hERG, the hERG/G572R mutant was generated by site-directed mutagenesis of the WT/hERG complementary DNA (cDNA) and then subcloned into the WT/hERG pcDNA3 vector at the BstEII/XhoI restriction site. The pcDNA3-WT and pcDNA3-G572R plasmids were transformed into Trans5α Chemically Competent Cell (cat. no. CD201-01, TransGen Biotech Co., Ltd.) and amplified. Expression constructs pcDNA3/hERG (WT)-FLAG and pcDNA3/hERG (G572R)-FLAG were engineered by ligating an oligonucleotide encoding a FLAG epitope to the carboxyl terminus of WT/hERG cDNA or hERG/G572R mutant cDNA. The pcDNA3-WT, pcDNA3-G572R, and pcDNA3-WT/pcDNA3-G572R were introduced into HEK293 cells by transient transfection using TransIT®-2020 Transfection Reagent (cat. no. MIR 5400; Mirus, Madison, WI, USA) to construct WT, mutant, and heterozygous cell lines, respectively.
According to the TransIT®-2020 Transfection Reagent instructions, a mixture of 2.5 µg of pcDNA3-WT, pcDNA3-G572R, or pcDNA3-WT/G572R plasmid, 250 µL Opti-MEM I Reduced Serum Medium (Gibco, Carlsbad, MA, USA), and 5 µL TransIT®-2020 Transfection Reagent was incubated for 20 minutes at room temperature and transferred to 293 cells containing 10% fetal bovine serum (FBS) medium. Subsequently, HEK293 cells were cultured in a 5% CO2 humidified cell incubator at 37 °C. The aim was to construct WT, hERG/G572R mutant, and WT/G572R heterozygous HEK293 cell models.
Western blot
At a quantity of 30 µg/lane, proteins were separated by electrophoresis on 7.5% sodium dodecyl sulfate (SDS) polyacrylamide gels and then transferred to 0.45 µm polyvinylidene fluoride (PVDF) membranes (Millipore, Burlington, MA, USA) by electroblotting. The PVDF membranes were blocked with blocking solution for 2 hours at room temperature to remove non-specific background. The primary antibody was diluted according to the instructions and the PVDF membranes were shaken overnight in tris-buffered saline with Tween 20 (TBST) containing the primary antibody. After washing the PVDF membranes in TBST 3 times for 5 minutes each time, secondary antibodies from the same species were added and incubated for 1 hour at room temperature. Visualization of PVDF membranes was performed using a WesternBright ECL WB detection kit (cat. no. K-12045-D10; Advansta, San Jose, CA, USA) under an ImageQuant LAS 500 imager (GE Healthcare, Chicago, IL, USA), and quantification of grayscale was conducted using ImageJ (National Institutes of Health, Bethesda, MD, USA). WB assay detected the protein expression of hERG (hERG/G572R, WT/G572R) before and after 4-PBA administration.
Quantitative real-time polymerase chain reaction
Total RNA was isolated from treated HEK293 cells using the TransZol Up Plus RNA Kit (cat. no. ER501-01; TransGen Biotech Co., Ltd.). TransScript® All-in-One First-Strand cDNA Synthesis SuperMix for qPCR (One-Step gDNA Removal) (cat. no. AT341-01; TransGen Biotech Co., Ltd.) reverse transcription was performed at 42 °C for 15 minutes and inactivated at 85 °C for 5 seconds. Reverse transcription turns RNA into cDNA. qPCR was performed using TransStart Tip Green qPCR SuperMix (+Dye II) (cat. no. AQ142-21, TransGen Biotech Co., Ltd.) and specific primers. The qPCR reaction procedure was as follows: 94 °C for 30 seconds, 94 °C for 5 seconds, and 60 °C for 30 seconds. Relative quantification was calculated using the 2−ΔΔCt method, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used to normalize mRNA expression. Primers for qPCR are shown in Table 1. qPCR assay detected the mRNA expression of hERG (WT/G572R) before and after 4-PBA administration.
Table 1
| Gene | Forward primer (5'–3') | Reverse primer (5'–3') |
|---|---|---|
| hERG | CAACCTGGGCGACCAGATAG | GGTGTTGGGAGAGACGTTGC |
| ATF6 | AGCAGCACCCAAGACTCAAAC | GCATAAGCGTTGGTACTGTCTGA |
| GRP78 | CATCACGCCGTCCTATGTCG | CGTCAAAGACCGTGTTCTCG |
| GRP94 | CCAGTTTGGTGTCGGTTTCTAT | CTGGGTATCGTTGTTGTGTTTTG |
| CRT | CCTGCCGTCTACTTCAAGGAG | GAACTTGCCGGAACTGAGAAC |
| CNX | GGCCAAGCATCATGCCATCT | TGATCCAGGTTGAGTTCTGGT |
| HRD1 | CTTCACCGTTTTTCGGGATGA | CCAGGAGGAACATAAGAGAGACA |
| GAPDH | ACAACTTTGGTATCGTGGAAGG | GCCATCACGCCACAGTTTC |
hERG, human ether-à-go-go-related gene; ATF6, activating transcription factor 6; CRT, calreticulin; CNX, calnexin; HRD1, 3-hydroxy-3-methylglutaryl coenzyme A reductase degradation protein 1; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Co-immunoprecipitation
HEK293 cells expressing hERG (WT/G572R) were harvested after 48 hours of transient transfection and 24 hours of treatment with 5 mM 4-PBA. Subsequently, the immunoprecipitation (IP)/co-IP kit (cat. no. abs955; Absin Biotechnology Co., Ltd., Shanghai, China) instructions were followed. The cells were lysed with the lysis buffer inside the kit and the supernatant collected after centrifugation. The lysate is used as the input group to determine the presence of the target protein in the cell lysate. Then, 5 µL protein A and 5 µL protein G were added to 500 µL of the cell lysate and left at 4 °C overnight. Phenylmethylsulfonyl fluoride (PMSF) and cocktail were added to the cell lysate every 3 hours to prevent protein degradation. Then, rabbit control IgG (cat. no. AC005, ABclonal, Wuhan, China) and anti-FLAG antibody (cat. no. ab205606, Abcam) were added as the IgG negative control group and FLAG positive control group, respectively. Thereafter, treatment was continued according to the IP/co-IP kit instructions. Protein samples (15–30 µL) were loaded onto 7.5% SDS-polyacrylamide gel electrophoresis (PAGE) gels and analyzed by WB. co-IP assay was used to discover the interaction of WT/G572R with ER molecular chaperone GRP78, GRP94, and ERAD genes HRD1.
Whole-cell patch-clamp electrophysiological recordings
WT/G572R heterozygous HEK293 cells from 4-PBA untreated and treated groups were collected, resuspended with trypsin, and incubated at 37 °C for 4 hours. Using a pipette with an end resistance of 2.0–6.0 MΩ, the capacitance and resistance of the whole cell was adjusted when the series resistance compensation reached 80%. Electrodes were connected to an Axopatch 700B amplifier, and currents were analog filtered at 2 kHz frequency and acquired and analyzed by a Digidata 1550 B digitizer and pCLAMP10.3 (Axon Instruments, Scottsdale, AZ, USA). Whole-cell patch-clamp electrophysiological recordings detected the channel current (IhERG) of hERG (WT/G572R) before and after 4-PBA administration.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 7.0 Software (GraphPad Software, Inc., San Diego, CA, USA), all experiments were performed at least 3 times, and data are presented as mean ± standard deviation (SD). The one- and two-way analysis of variance (ANOVA) with Dunnett’s post-hoc test or two-tailed Student’s t-test was used. For electrophysiological normalized data analysis, a Wilcoxon Mann-Whitney U test was used to compare currents with the drug to control currents. Significance was calculated at a 95% confidence interval (CI), and P<0.05 (two-sided) was considered a statistically significant difference.
Results
4-PBA reversed the electrophysiological phenotype of the WT/G572R channel
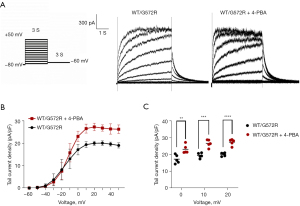
The hERG (WT/G572R) current (IhERG) was recorded in cells grown without the drug and in cells grown after incubation with 5 mM 4-PBA. As shown in Figure 2A, whole-cell patch-clamp currents were observed in WT/G572R cells, indicating that heterozygous WT/G572R was not completely functionally lost, but WT/G572R whole-cell patch-clamp currents were more significant after 5 mM drug treatment. In Figure 2B, I–V curves of tail current density (pA/pF) in WT/G572R and WT/G572R + 4-PBA, normalized data for tail currents of WT/G572R and WT/G572R + 4-PBA are plotted against the test potential and fitted to Boltzmann functions. In addition, the results suggested that the hERG (WT/G572R) tail current was substantially increased after drug treatment (17.201 vs. 23.133 in 0 mV, P=0.0070; 19.428 vs. 26.647 in 10 mV, P=0.0003; 20.061 vs. 27.375 in 20 mV, P<0.0001) (Figure 2B,2C).
hERG/G572R expression was increased by 4-PBA
To better understand the effect of 4-PBA on hERG/G572R channel protein maturation and trafficking, wild, mutant, and heterozygous cell lines without 4-PBA treatment were used as the control group, and mutant and heterozygous cell lines treated with 5 mM 4-PBA were used as the experimental group. As shown in Figure 3A, the first 3 lanes represent WT, mutant (G572R), and heterozygous (WT/G572R) controls without 4-PBA treatment, and the fourth and fifth lanes represent mutant and heterozygous experimental groups, respectively, with 4-PBA treatment. In the hERG lane, there are 2 proteins at the top and the bottom, with the top 155 kDa hERG representing the mature form of fully glycosylated in the cell membrane and the bottom 135 kDa hERG representing the immature form of core glycosylated in the ER.
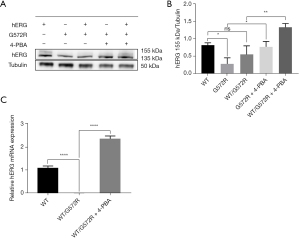
As displayed in Figure 3A, compared with the WT/hERG with normal function, we found that the mutant and heterozygous control group without 4-PBA treatment had a significant reduction in 155 kDa, indicating that the hERG/G572R protein was retained in the ER and had a transport defect, indicating that WT/G572R had a clear dominant negative inhibition effect. As shown in Figure 3B, we also found that the functional 155 kDa in the cell membrane of the mutant and heterozygous experimental group after the 4-PBA treatment was significantly more than that of the mutant (0.2873 vs. 0.7807, P=0.0269) and heterozygous control group (0.5597 vs. 1.3450, P=0.0011) without the 4-PBA treatment. This suggests that 4-PBA can correct the hERG/G572R channel trafficking defect. From Figure 3C, we can infer that heterozygous WT/G572R may have nuclear transcriptional abnormalities (3). As it happens, 4-PBA is not only an ERS inhibitor but also a transcriptional regulator. Relative to controls without 4-PBA treatment, 5 mM 4-PBA increased WT/G572R mRNA expression (0.01662 vs. 2.376, P<0.0001) in the nucleus as a result of more DNA transcribed to produce RNA.
4-PBA inhibited the ERS
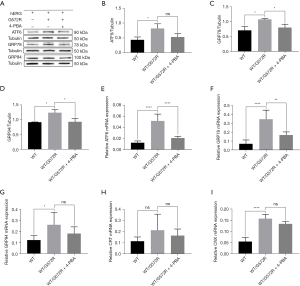
In general, mutations are manifestations of heterozygous mutations. To better understand the mechanism by which the ERS inhibitor 4-PBA exerts its effect on the hERG/G572R channel, we tested the expression levels of markers of ERS, GRP94, GRP78, CRT/CNX, and ATF6 pathway in UPR before and after 4-PBA treatment in WT/G572R heterozygous cell lines. As shown in Figure 4A-4D, in the heterozygous WT/G572R cell line, ATF6 [0.8160 vs. 0.5268, non-significant (ns)], GRP78 (1.081 vs. 0.8079, P=0.0373), and GRP94 (1.2320 vs. 0.9250, P=0.0144) also decreased at the protein level after applying 5 mM 4-PBA. As shown in Figure 4E-4I, compared with the WT/hERG, the heterozygous WT/G572R resulted in an mRNA up-regulation of ATF6 (0.01248 vs. 0.05161, P<0.0001; 0.05161 vs. 0.02098, P<0.0001), GRP78 (0.06956 vs. 0.3444, P<0.0001; 0.3444 vs. 0.1684, P=0.0034), GRP94 (0.1227 vs. 0.2589, P=0.0397; 0.2589 vs. 0.1808, ns), CRT (0.1149 vs. 0.2125, ns; 0.2125 vs. 0.1635, ns) and CNX (0.05440 vs. 0.1569, P<0.0001; 0.1569 vs. 0.1338, ns), all of which were down-regulated after treatment with 5 mM 4-PBA for 24 hours. These findings reveal that WT/G572R induces ERS responses via the ATF6 pathway, whereas 4-PBA inhibits the expression of the ATF6 pathway and the expression of related molecular chaperones.
4-PBA inhibited the HRD1 gene downstream of ERS
The point mutation hERG/G572R channel induces UPR and ERS response. ERS maintains homeostasis in the cell and ERAD degrades excess unfolded proteins, and E3 ubiquitin ligase HRD1 is one of the key genes in ERAD. As shown in Figure 5A-5C, heterozygous WT/G572R of Mendelian genetic disease alleles caused a significant increase in E3 ubiquitin ligase HRD1 compared with WT, and HRD1 decreased at both mRNA (0.001685 vs. 0.0008124, P=0.0001) and protein levels (1.264 vs. 0.9836, P=0.0062) after 4-PBA treatment.
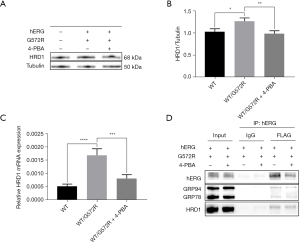
HEK293 cell lysates expressing WT/G572R, and WT/G572R + 4-PBA were immunoprecipitated with anti-hERG FLAG antibodies and immunoblotted with anti-GRP78 antibodies, anti-GRP94 antibodies, and anti-HRD1 antibodies. As shown in Figure 5D, lanes 1 and 2 represent whole cell lysates of WT/G572R and WT/G572R + 4-PBA. It can be seen that GRP78, GRP94, and HRD1 target proteins do exist in cell lysates. Lanes 3 and 4 represent the IgG negative control group of WT/G572R and WT/G572R + 4-PBA, respectively, excluding false positives caused by operation or reagents. Lane 5 and lane 6 represent the positive experimental group of WT/G572R and WT/G572R + 4-PBA. Our data suggest that WT/G572R can indeed interact with the ERAD gene E3 ubiquitin ligase HRD1 and that WT/G572R also interacts with the ER molecular chaperones GRP78 and GRP94.
Discussion
In this study, our findings suggest that ERS inhibitor 4-PBA can inhibit ERS, attenuate the UPR, and further increase the membrane localization and IhERG of the WT/G572R channel. By attenuating the UPR, 4-PBA allows more misfolded proteins to be exported from the ER and is less susceptible to degradation by the ERAD pathway. The hERG/G572R mutation causes an ERS response. Classical ERS agonists include tunicamycin, which inhibits protein glycosylation, and thapsigargin, an ER calcium ATPase inhibitor. ERS inhibitors include ursodeoxycholic and tauroursodeoxycholic acid (UDCA and TUDCA), taurine (Tau), and salburinal (Sal), among others. A previous study has reported that chemical and pharmacological chaperones, such as TUDCA, reduced degradation and/or promoted plasma-membrane localization of defective subunits in cyclic nucleotide-gated (CNG) channel mutations (13). Whether the ERS inhibitors UDCA, TUDCA, Tau, and Sal can correct the transport/plasma-membrane localization defect in the hERG/G572R channel remains to be investigated.
Potential molecular chaperone in hERG/G572R channel
Molecular chaperones exist in mitochondria, ER, cytoplasm, and other intracellular parts. Potential molecular chaperones for the hERG/G572R channel include a family of heat shock proteins (HSPs), the nucleoplasmin family (Npm1, Npm2a, Npm2b, Npm3), and the TCP-1 family (TRiC, CCT1-8). HSPs include small heat shock proteins (sHsps), heat shock proteins 40s/J domain proteins (Hsp40s), heat shock proteins/chaperonin 60s (Hsp60s), heat shock protein 70s (Hsp70s), heat shock protein 90s (Hsp90s), heat shock protein 100s (Hsp100s), and large stress/HSPs (Hsp110, Grp170). Protein disulfide isomerase (PDI) and peptidyl-prolyl cis-trans isomerase (PPI) are the most important members of folding assist enzymes, chaperones that contribute to the folding of ER proteins in ERS, and important potential therapeutic targets of hERG/G572R channel mutations (14). It has not been clarified whether overexpression of nuclear chaperones (Npm1, Npm2a, Npm2b, Npm3) can increase transcription of hERG/G572R mRNA in the nucleus. Hsp90 can be considered a “molecular buffer” that maintains the overall cell balance; chaperonin (HSPs, TRiC) may be involved in the pathogenesis of many diseases and even become biomarkers for the risk and prognosis of some diseases. Hsp90 inhibitors are being investigated for the treatment of various diseases, including cancer therapy and the treatment of many inflammatory diseases. It has even been reported that the Hsp70 blocker pifithrin-µ or the Hsp90 blocker 17-DMAG can restore mature glycosylation in some of the CTD-causing hCRT-1 mutants (11). The chaperone protein can be available on the GeneCards website. Potential molecular chaperones of the hERG/G572R channel are presented in Table 2, which may be new approaches to finding treatments for inherited genetic mutation diseases.
Table 2
| Categories | Gene symbols |
|---|---|
| NPM family | Npm1, Npm2a, Npm2b, Npm3 |
| TCP-1 family | TRiC (CCT1-8) |
| HSPs | |
| sHsps (12–34 kDa) | HSPB1/HSP27/HSP28/Hsp25, HSPB8/Hsp22, Hsp32 |
| Hsp40s (40 kDa) | DnaJ/Hsp40 (ERdj1, ERdj3, ERdj4, ERdj6) |
| Hsp60s (60 kDa) | Hsp60 (GroEL) |
| Hsp70s (70 kDa) | HSP75 (GRP75), HSP70 (HSP72), HSPA8 (HSC70/HSC71/HSP73) |
| Hsp90s (90 kDa) | gp96/ERp99 |
| Hsp100S (100 kDa) | ClpA, ClpB/Hsp104, ClpC, ClpX, HslU (15) |
| HSPHs (110 kDa) | Hsp110s (Hsp105, HspA4, HspA4L) |
hERG, human ether-à-go-go-related gene; NPM, nucleoplasmin; HSPs, heat shock proteins; sHsps, small heat shock proteins.
Potential ERAD genes in hERG/G572R channel
ER can sense certain stimuli inside and outside the cell, and misfolded, unfolded, and unassembled proteins can be degraded through the ERAD pathway, which is one of the key mechanisms for maintaining homeostasis. ERAD includes the ubiquitin-proteasome pathway (UPP) and autophagy-lysosomal pathway (ALP). UPP includes ubiquitin, E1 ubiquitin-activating enzyme (UBA), E2 ubiquitin-conjugating enzyme (UBC), E3 ubiquitin-ligating enzymes, 26S proteasomes, and deubiquitinating enzymes (DUBs). E3 ubiquitin ligases mainly include the RING finger domain (RING), homologous to E6-associated protein C-terminus (HECT), U-box domain (U-box), and Cullin-RING ligases (CRLs). DUBs are divided into 6 main subcategories: ubiquitin-specific proteases (USPs or UBPs), ubiquitin C-terminal hydrolases (UCHs), Machado-Joseph disease proteases (MJDs), Ovarian tumor domain proteases (OTUs), JAB1/MPN/Mov34 metalloenzymes (JAMMs), and monocyte chemotactic protein-induced proteins (MCPIPs). ALPs include macroautophagy (MA), microautophagy (MI), and chaperone-mediated autophagy (CMA). E3 ubiquitin ligase is a core component of ERAD, and the 26S proteasome plays a key role in ubiquitinated protein degradation and is responsible for the degradation of most cellular proteins; it is also the executive arm of the UPP (16). The potential ERAD genes of the hERG/G572R channel include derlin1/2/3, P97/VCP, CHIP, and others. The proteasome inhibitor lactacystin enhances the level of mature 155 kDa hERG in the N996I/LQTS2 mutation, and the broad-spectrum ubiquitin-proteasome inhibitor ALLN reverses the gating kinetics of hERG/A561V (17). Many UPP inhibitors such as bortezomib, carfilzomib, and ixazomib have been comprehensively used in cancer therapy, and UPP has been developed as a promising therapeutic target for novel anticancer agents. Table 3 illustrates the potential ERAD genes of the hERG/G572R channel, which may be a new direction for finding the treatment of inherited gene mutation diseases.
Table 3
| Categories | Gene symbols |
|---|---|
| UPP | |
| Ub | UBQLN1-4, UBQLNL |
| E1s | UBA1/UBE1, UBA6/UBE1L2 |
| E2s | UBE2N, UBE2L3, UBE2D2/3, UBE2C |
| E3s | RING (Pellino, UBR1, RNF5/8/19/190/20/34/40/125/128/138/168, TMEM129); HECT (HERC E3s, NEDD4-2); U-box (CHIP); CRLs (CRL1/2/3/4/5/6/7) |
| 26S proteasomes (CP, RP) | Rpns |
| DUBs | USPs (USP52, USP25, USP19, USP15, USP13, USP14); UCHs (Uch37/UchL5); MJDs; OTUs (OTULIN, YOD1); JAMMs; MCPIPs |
| ALP | |
| MA | LC3 (LC3B) |
| MI | eMI |
| CMA | HSP70, LAMP2A |
ERAD, endoplasmic reticulum-associated degradation; UPR, unfolded protein response; hERG, human ether-à-go-go-related gene; UPP, ubiquitin-proteasome pathway; Ub, ubiquitin; ALP, autophagy-lysosomal pathway; CP, 20S core particle; RP, 19S regulatory particles; DUBs, deubiquitinating enzymes; MA, macroautophagy; MI, microautophagy; CMA, chaperon-mediated autophagy; HECT, homologous to E6-associated protein C-terminus; U-box, U-box domain; CRLs, cullin-RING ligases; USPs, ubiquitin-specific proteases; UCHs, ubiquitin C-terminal hydrolases; MJDs, Machado-Joseph disease proteases; OTUs, ovarian tumor domain proteases; JAMMs, JAB1/MPN/Mov34 metalloenzymes; MCPIPs, monocyte chemotactic protein-induced proteins; eMI, endosomal microautophagy.
Potential apoptotic genes in hERG/G572R channel
If the body is subjected to persistent severe ERS, the ERS will activate apoptotic signals. Apoptotic genes include anti-apoptotic genes and pro-apoptotic genes. CHOP (C/EBP homologous protein, also known as GADD153) is the common pro-apoptotic gene, affecting the eIF2α-ATF4/ATF6-CHOP-Bim/Bax signaling pathway. It is also highly likely that CHOP is an apoptotic gene downstream of hERG/G572R. Pro-apoptotic genes mainly include the cysteinyl aspartate specific proteinase (Caspase) family, interleukin-1 beta-converting enzyme (ICE) family, apoptotic protease activating factor-1 (Apaf-1), tumor necrosis factor receptor (TNFR) family, p53-related nuclear transcription factors family, and MYC family. Apoptosis inhibitors include broad-spectrum apoptosis inhibitor P35, cytokine response modifier A (CrmA), and other inhibitors of apoptosis proteins (IAPs), flice-like inhibitory proteins (FLIPs), NOD-like receptor (NLR) family, and B-cell leukemia/lymphoma-2 (Bcl-2) family. The Bcl-2 family has both anti-apoptotic and pro-apoptotic effects. Genistein (GS) plays a neuroprotective role in Alzheimer’s disease (AD) by inhibiting the ER-mediated apoptotic pathway (18), and the anti-apoptotic gene survivin is a therapeutic target in malignant rhabdoid tumor (MRT) and other SMARCB1/INI1-deficient tumors (19-21). Table 4 illustrates the various potential apoptotic genes of the hERG/G572R channel, which may be a new way to find the treatment of inherited gene mutation diseases.
Table 4
| Categories | Gene symbols |
|---|---|
| Pro-apoptosis | Caspase family (Caspase 3, Caspase 6, Caspase 7, Caspase 8, Caspase 9); ICE family (caspase-1); Apaf-1; TNFR family (Fas/CD95, TNFR1, DR3/WSL-1, DR4/TRAIL-R1, DR5/TRAIL-R2); p53 family (p53/TP53, p63/TP63, p73/TP73); MYC family (c-Myc); Bcl-2 family (Bok, Bax, Bak, Bad, Bid, Bim, Bmf, Bcl-xs, Mtd, Bik/Nbk, Harakiri, BNIP3L, Noxa) |
| Anti-apoptosis | Bcl-2 family (Bcl-2, Bcl-xl, Bcl-w, Mcl-1, A1, ced-9); P35; IAPs (XIAP, cIAP-1/2, Livin, Survivin, BRUCE); CrmA; FLIPs; NLR family (NAIP) |
hERG, human ether-à-go-go-related gene; ICE, interleukin-1 beta-converting enzyme; Apaf-1, apoptotic protease activating factor-1; TNFR, tumor necrosis factor receptor; Bcl-2, B-cell leukemia/lymphoma-2; IAPs, inhibitors of apoptosis proteins; CrmA, cytokine response modifier A; FLIPs, flice-like inhibitory proteins; NLR, nucleotide-binding and oligomerization domain (NOD)-like receptor.
Conclusions
Our work provides preliminary evidence that 4-PBA corrects hERG channel transport defects by inhibiting excessive ERS and ERAD-related gene E3 ubiquitin ligase HRD1. Additionally, 4-PBA increased WT/G572R mature functional channel protein and improved channel current. 4-PBA is expected to be developed as a new treatment method for LQT2 (WT/G572R). But more large-scale multicenter trials are needed to verify it. With the deepening of research, further understanding of 4-PBA will provide new ideas for treating LQT2.
Acknowledgments
Funding: This study was supported by the National Natural Science Foundation of China (No. 81870255 to JL), Zhejiang Provincial Science Foundation of China (Nos. LY21H020001 to JL and LQQ20H160001 to YS), Zhejiang Provincial Medicine & Healthcare Technology Project of China (No. 2021KY306 to JL), and Ningbo Municipal Science Foundation of China (No. 2021J296 to YS).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1252/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1252/dss
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1252/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1252/coif). YS reports that this study was supported by Ningbo Municipal Science Foundation of China (No. 2021J296 to YS), and Zhejiang Provincial Science Foundation of China (No. LQQ20H160001 to YS). JL reports that this study was supported by the National Natural Science Foundation of China (No. 81870255 to JL), Zhejiang Provincial Science Foundation of China (No. LY21H020001 to JL), Zhejiang Provincial Medicine & Healthcare Technology Project of China (No. 2021KY306 to JL). The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ono M, Burgess DE, Schroder EA, et al. Long QT Syndrome Type 2: Emerging Strategies for Correcting Class 2 KCNH2 (hERG) Mutations and Identifying New Patients. Biomolecules 2020;10:1144. [Crossref] [PubMed]
- Vandenberg JI, Perry MD, Perrin MJ, et al. hERG K(+) channels: structure, function, and clinical significance. Physiol Rev 2012;92:1393-478. [Crossref] [PubMed]
- Anderson CL, Kuzmicki CE, Childs RR, et al. Large-scale mutational analysis of Kv11.1 reveals molecular insights into type 2 long QT syndrome. Nat Commun 2014;5:5535. [Crossref] [PubMed]
- Zhang Y, Cen J, Jia Z, et al. Hepatotoxicity Induced by Isoniazid-Lipopolysaccharide through Endoplasmic Reticulum Stress, Autophagy, and Apoptosis Pathways in Zebrafish. Antimicrob Agents Chemother 2019;63:e01639-18. [Crossref] [PubMed]
- Rani S, Sreenivasaiah PK, Kim JO, et al. Tauroursodeoxycholic acid (TUDCA) attenuates pressure overload-induced cardiac remodeling by reducing endoplasmic reticulum stress. PLoS One 2017;12:e0176071. [Crossref] [PubMed]
- Shruthi K, Reddy SS, Chitra PS, et al. Ubiquitin-proteasome system and ER stress in the brain of diabetic rats. J Cell Biochem 2019;120:5962-73. [Crossref] [PubMed]
- Gong J, Fang L, Liu R, et al. UPR decreases CD226 ligand CD155 expression and sensitivity to NK cell-mediated cytotoxicity in hepatoma cells. Eur J Immunol 2014;44:3758-67. [Crossref] [PubMed]
- Kaneko M, Nomura Y. ER signaling in unfolded protein response. Life Sci 2003;74:199-205. [Crossref] [PubMed]
- You YD, Deng WH, Guo WY, et al. 4-Phenylbutyric Acid Attenuates Endoplasmic Reticulum Stress-Mediated Intestinal Epithelial Cell Apoptosis in Rats with Severe Acute Pancreatitis. Dig Dis Sci 2019;64:1535-47. [Crossref] [PubMed]
- Zhou X, Xu Y, Gu Y, et al. 4-Phenylbutyric acid protects islet β cell against cellular damage induced by glucocorticoids. Mol Biol Rep 2021;48:1659-65. [Crossref] [PubMed]
- El-Kasaby A, Kasture A, Koban F, et al. Rescue by 4-phenylbutyrate of several misfolded creatine transporter-1 variants linked to the creatine transporter deficiency syndrome. Neuropharmacology 2019;161:107572. [Crossref] [PubMed]
- Tominaga K, Tominaga N, Williams EO, et al. 4-Phenylbutyrate restores localization and membrane repair to human dysferlin mutations. iScience 2022;25:103667. [Crossref] [PubMed]
- Duricka DL, Brown RL, Varnum MD. Defective trafficking of cone photoreceptor CNG channels induces the unfolded protein response and ER-stress-associated cell death. Biochem J 2012;441:685-96. [Crossref] [PubMed]
- Hirsch I, Weiwad M, Prell E, et al. ERp29 deficiency affects sensitivity to apoptosis via impairment of the ATF6-CHOP pathway of stress response. Apoptosis 2014;19:801-15. [Crossref] [PubMed]
- Hodson S, Marshall JJ, Burston SG. Mapping the road to recovery: the ClpB/Hsp104 molecular chaperone. J Struct Biol 2012;179:161-71. [Crossref] [PubMed]
- Pathare GR, Nagy I, Bohn S, et al. The proteasomal subunit Rpn6 is a molecular clamp holding the core and regulatory subcomplexes together. Proc Natl Acad Sci U S A 2012;109:149-54. [Crossref] [PubMed]
- Mehta A, Sequiera GL, Ramachandra CJ, et al. Re-trafficking of hERG reverses long QT syndrome 2 phenotype in human iPS-derived cardiomyocytes. Cardiovasc Res 2014;102:497-506. [Crossref] [PubMed]
- Gao H, Lei X, Ye S, et al. Genistein attenuates memory impairment in Alzheimer's disease via ERS-mediated apoptotic pathway in vivo and in vitro. J Nutr Biochem 2022;109:109118. [Crossref] [PubMed]
- Yoshino Y, Goto H, Ito M, et al. YM155 and chrysin cooperatively suppress survivin expression in SMARCB1/INI1-deficient tumor cells. Med Oncol 2022;39:234. [Crossref] [PubMed]
- Zhang G, Wang B, Cheng S, et al. KDELR2 knockdown synergizes with temozolomide to induce glioma cell apoptosis through the CHOP and JNK/p38 pathways. Transl Cancer Res 2021;10:3491-506. [Crossref] [PubMed]
- Lin L, Zhang W. Development and validation of endoplasmic reticulum stress-related eight-gene signature for predicting the overall survival of lung adenocarcinoma. Transl Cancer Res 2022;11:1909-24. [Crossref] [PubMed]


