
Distribution of common and rare drug resistance patterns in Mycobacterium tuberculosis clinical isolates revealed by GenoType MTBDRplus and MTBDRsl assay
Highlight box
Key findings
• The most prevalent mutations observed were rpoB S531L and inhA promoter C15T.
What is known and what is new?
• The epidemiology, transmission, genotypes, mutational patterns, and clinical consequences of tuberculosis (TB) have been extensively studied worldwide, there is a lack of information regarding the epidemiology and mutation patterns of Mtb in Pakistan, specifically concerning the prevalence of multi-drug resistant TB.
• The current study added new information on epidemiology and mutational patterns of Mycobacterium tuberculosis (Mtb) in Pakistan.
What is the implication, and what should change now?
• The findings contribute to our understanding of multi-drug resistant-TB epidemiology and provide insights into the genetic profiles of Mtb in Pakistan, which are essential for effective TB control strategies.
Introduction
Drug-resistant tuberculosis (DR-TB) has become a serious public health threat due to an escalation in the incidences worldwide, which needs immediate attention. DR-TB leads to multi-DR-TB (MDR-TB), when the Mycobacterium tuberculosis (Mtb) develops resistance to the two most important first-line anti-tuberculosis drugs (FLDs), rifampicin (RIF) and isoniazid (INH), either with or without any additional FLDs. These MDR-TB strains develop due to ignorance, illiteracy (less awareness about TB), poor compliance, improper treatment or poor management of drug-susceptible tuberculosis (DS-TB). Although some of these causes may vary by region/country, generally they are the same, thus constituting a major impediment to the fight against tuberculosis (TB) and posing an imminent threat to the world (1).
According to the WHO 2022 report, in 2021 there were an estimated 10.6 million new and previously treated cases of Mtb complex (MTBC) infections, including 450,000 cases of MDR-TB and RIF-resistant TB (RR-TB), indicating a high prevalence of MDR-TB (2). Despite significant medical advancements and social interventions, TB management remains challenging due to the potential for hidden transmission. Infected individuals can unknowingly spread the disease to others, as the progression from latent TB infection to active TB disease can occur gradually over time. Also, in most cases, the infected patients remain undiagnosed, posing a significant risk of widespread transmission of infection (3). The presence of undiagnosed rifampicin-resistant (RIFR) cases has exacerbated the global health situation and contributed to the escalating burden of TB (4). Furthermore, public health statistics have indicated that the cost of treating DR-TB is 20-fold higher than that of DS-TB (5).
On one hand, the treatment duration for DR-TB varies widely, ranging from the recently introduced short-term regimens to longer-term regimens lasting between 9 and 24 months. Additionally, second-line anti-TB drugs (SLDs) used in the treatment of DR-TB pose challenges due to their limited availability, high cost, and associated issues compared to FLDs. On the other hand, SLDs have been associated with unfavorable clinical outcomes, including significant adverse pharmacological effects. Furthermore, numerous trials have reported therapeutic success rates of only 60% to 70% (5), thereby intensifying the difficulty in combating the disease. Also, other kinds of treatment include regimens containing newer drugs, adjunctive therapies (immunomodulatory drugs, host-directed therapies, and nutritional supplementation to support the patient’s immune system), and other medicines such as traditional Chinese medicine and even surgery are alternative treatment options and may vary depending on the specific healthcare setting, drug accessibility, and local guidelines. The choice of treatment approach is typically made by healthcare professionals based on individual patient factors, drug resistance patterns, and the expertise available (6,7).
In the context of TB, Pakistan ranks alongside India, China and the Russian Federation as having a high TB burden. In Pakistan, the incidence of TB is 268 per 100,000 individuals, and the incidence of MDR-TB is 14 per 100,000 population (2). Studies on the prevalence of DR-TB, including MDR-TB and extreme DR-TB (XDR-TB), are minimal and inconsistent (6). Thus, data acquired through national surveys on DR-TB and reported to WHO must be viewed with caution. Also, the actual prevalence of DR-TB is thought to be underestimated owing to scarcity of facilities that offer drug susceptibility testing (DST) and lack of current and advanced technologies that allow for quick and accurate drug resistance profiling (7).
Thus the problem with global TB statistics, especially in Pakistan, warrants rigorous case report monitoring, accurate diagnosis and follow-up through effective national surveillance programs. For instance, the primary method for diagnosing TB is sputum smear microscopy. Recently, however, molecular methods like GeneXpert, polymerase chain reaction (PCR), and line probe assays (LPAs) were employed for the quick identification of DR-TB to ensure adequate treatment and track TB load. Therefore, the WHO recommends LPAs for rapid diagnosis of patients with risk of MDR-TB (4). The common LPA such as GenoType MTBDRplus assay is used for both clinical samples and culture isolates to detect rpoB, katG and promoter region of inhA, while MTBDRsl is used to detect mutation in SLDs [levofloxacin (LFX) and moxifloxacin (MOX)]. Also, the MTBDRplus assay helps in the detection of a high or low-level resistance to INH by the katG and inhA genes, and the MTBDRsl helps in the detection of rpoB mutation of fluoroquinolones (FQs) (8). The current study aims to compare the utility of LPA in our setup to solid and automated liquid culture systems and to establish the prevalence of primary drug resistance within the study population. We present this article in accordance with the STROBE reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-138/rc).
Methods
The study participants were selected based on specific demographic and clinical characteristics. Demographic information such as age, gender, and geographical location was collected for each patient. Clinical characteristics, including the TB treatment history, were also recorded. In addition, patients were categorized into subgroups based on their treatment status, distinguishing between new and retreated cases. Furthermore, strains were classified as belonging to the Beijing family (n=191) or non-Beijing strains (n=50) based on genotyping methods, such as PCR.
Ethical approval
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the ethics committee of Saidu Teaching Hospital (DRTB/NTP/022), and informed consent was taken from all the patients.
Inclusion and exclusion criteria
The samples were included on the basis of confirmed TB-diagnosed cases, newly diagnosed or previously treated TB cases, geographical location, time period and drug resistance. While others were excluded based on extrapulmonary TB, co-infections or comorbidities, insufficient data, and duplicate cases. Also, samples which were not resistant on both GenXpert and LPA methods were excluded.
Sample assembly and storage
Over a one-year time span, we collected early-morning sputum samples in sterile leak-proof containers from 241 patients who were smear positive with a grade of +1 or higher. The samples were kept in freezer at temperature of 2–8 ℃ for a maximum of four days.
Specimen processing
The Ziehl Neelsen procedure was used to stain the specimens. For 1–10 acid-fast bacilli (AFB)/100 oil emulsion fields (OIF), the smears were evaluated as “scanty”, “1+” for 10–99 AFB/100 OIF, “2+” for 1–9 AFB/OIF, and “3+” for >10 AFB/OIF. Smears that were found to be “scanty” positive were not included in this investigation, and three replicates were performed for each case.
Decontamination
The decontamination procedure was carried out by using the N-acetyl-L-cysteine (NALC)-NaOH-sodium citrate method. For mixture preparation the 0.5 g NALC/100 mL of NaOH-sodium citrate solution was mixed with equal amount of NaOH (4%), and sodium citrate (2.9%), followed by the addition of sample and NALC into a 50 mL plastic sterile centrifuge tube. The tube was vortexed for around 20 s, and then allowed to stand for 20 min. The sterile phosphate buffer solution of pH 6.8 was added to tubes up to 50 mL mark, and mixed by hand spun. Then samples were concentrated by 3,000 g speed centrifuge for 15 min, and supernatant was discarded. Finally, to achieve a volume of 1 mL, the pellet was suspended by phosphate buffer (pH 6.8).
GeneXpert MTB/RIF assay
For early detection and appropriate treatment, the smear positive samples were subjected to the WHO-recommended assay. GeneXpert MTB/RIF which is used to rapidly identify the RIFR in MTBC (9). We investigated the clinical isolates using the GeneXpert MTB/RIF test, and followed the manufacturer’s methodology to detect the resistance-conferring mutations found in rifampicin-resistant determinant region (RRDR) of rpoB in Mtb. After determining the RIF-resistant isolates, the samples were subjected to the phenotypic DST.
DST of samples by MGIT 960 system
Following the GeneXpert assay, the clinical samples’ resistance to other antibiotics was assessed using the Bactec MGIT 960 system at the MDR-TB Center, Saidu Teaching Hospital Swat, Pakistan, in accordance with the manufacturer’s instructions and approved guidelines (4). According to WHO standards, phenotypic DST for RIF, INH, and FQ was carried out on the Bactec MGIT 960 system using 1, 0.1, and 1 µg/mL, respectively as the critical concentration (CC) (10). The wild-type Mtb H37Rv strain (ATCC27294T) was used as a control.
DNA extraction and amplification
For DNA extraction, 500 µL of decontaminated sample was loaded in 1.5 mL tubes and centrifuged at 10,000 g for 15 min. After centrifugation, the pellet was suspended in 200 µL of distilled water and incubated in a water bath at 95 ℃ for 20 min, followed by incubation for 15 min in an ultrasonic bath. This was spun down for 5 min at 14,000 g followed by final extraction of the DNA.
LPA hybridization
The Hain protocol was used for hybridization according to manufacturer’s instruction. The smear positive samples were incubated in a “Twin-Cubator”, a semiautomatic washing and shaking apparatus that can accommodate 12 samples at once. The hybridization process was performed by using the reagents provided with the kits. According to instruction, in each of the wells, the 20 mL of denaturation solution was distributed and amplification samples were then added and mixed carefully. Then hybridization buffer was added after 5 min of incubation, and a single strip was inserted in each well with the coated side facing up.
Following the incubation and washing operations, the diluted conjugate was added and stored for further incubation. The strips were then incubated in a darkened area after the substrate was added. For the washing process, the kit protocol of stringent wash was followed. The strips were removed with tweezers after a final wash with distilled water and dried between two layers of absorbent paper. The sheets were aligned with the corresponding lines on the evaluation sheet provided with the kit, and the bands CC and amplification control (AC) were inserted in the specified fields. Each strip contains 27 reaction zones and sensitivity is defined by the presence of wild-type bands without the presence of a mutant band, whereas resistance is defined by the appearance of a mutant band plus/minus a corresponding wild-type band.
All of the patients were registered with the MDR-TB Center at Saidu Teaching Hospital and were being followed up on a regular basis. Patients with smear-positive TB were started on a category I “directly observed treatment short-course” regimen, whereas those with confirmed MDR-TB were started on a category IV regimen.
Statistical analysis
The data interpretation and statistical summary were conducted using the SPSS version 16 and GraphPad Prism v5. All the variable outcomes were presented as percentages (%), and odds ratio (OR). Fisher’s exact test and Chi-Square test were employed to assess the associations between categorical variables. The results were reported as either mean values or the number of patients. Statistical significance was defined as a P value less than 0.05.
Results
In the present study, patients suspected to be infected with TB were studied for infection with DS- or DR-TB. A total of 1,303 suspected cases were studied, and of all the cases, 241 (18%) and 1,062 (82%) tested positive and negative respectively. Of the TB-positive cases, 112 (46%) were males and 129 (54%) were females. The proportion of positive cases was highest in the age group 15–24 years (n=84, 35%) followed by 25–34 (n=53, 22%) (Table 1). Based on the patients’ treatment history, new cases constitute more than half (n=126, 52%) of the cases, whereas the remaining (n=115, 48%) constitute the number of retreated cases.
Table 1
| Parameters | Sub-categories | Frequency | Percentage |
|---|---|---|---|
| Total cases | Positive | 241 | 18 |
| Negative | 1,062 | 82 | |
| Positive cases gender-wise distribution |
Male | 112 | 46 |
| Female | 129 | 54 | |
| Positive cases age-wise distribution (years) |
0–14 | 13 | 5 |
| 15–24 | 84 | 35 | |
| 25–34 | 53 | 22 | |
| 35–44 | 39 | 16 | |
| 45–54 | 21 | 9 | |
| 55–64 | 17 | 7 | |
| 65+ | 14 | 6 | |
| Positive cases treatment history-wise distribution | New cases | 126 | 52 |
| Retreated cases | 115 | 48 |
There were 97 incidences of mono-resistance among the 241 positive TB patients, and they were classified as DR. MDR accounted for 84 instances, followed by Pre-XDR, which accounted for 57 cases. However, we found a low rate of XDR, with only 3 incidents (Figure 1).
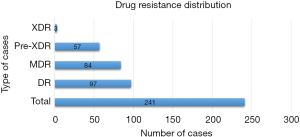
It was determined that rpoB is almost the unique gene for RIF resistance. The mutation S351L in the rpoB gene was found in 107 of the 241 TB-positive patients, followed by 40 instances with the mutation L530M in rpoB, and then the additional mutations listed in Table 2. In the 241 TB-positive patients, katG and inhA were responsible for INH resistance, with 63 and 74 cases respectively. At the same time, FQ resistance was seen to result from mutations in gyrA. The mutation D94Y was found in 24 of the FQ-resistant (FQR) isolates, followed by D94A (n=21), D94G (n=10) and D94N (n=6) in gyrA.
Table 2
| Drug | MIC (µg/mL) | Gene | Amino acid changed | Codon changed | MTBDRplus/MTBDRsl | No. of isolates |
|---|---|---|---|---|---|---|
| RIF | 1.0–160 | rpoB | S531L | TCG→TTG | Mut3 | 107 |
| L533P | CTG→CCG | Wt8 | 37 | |||
| L530M | CTG→CCG | Wt8 | 40 | |||
| H526Y | CAC→TAC | Mut2a | 21 | |||
| H526D | CAC→GAC | Mut2b | 22 | |||
| INH | 0.1 | katG | S315T | AGC→ACC | Mut1 | 63 |
| inhA | c-15t | Promoter up mutation | Mut1 | 74 | ||
| FQ | 1.0 | gyrA | D94A | GAC→GCC | Mut3a | 21 |
| D94G | GAC→GGC | Mut3c | 10 | |||
| D94N | GAC→AAC | Mut3b | 6 | |||
| D94Y | GAC→TAC | Mut3b | 24 |
MIC, minimum inhibitory concentration; RIF, rifampicin; INH, isoniazid; FQ, fluoroquinolone.
In the current study, Mtb isolates from TB-positive cases were studied for drug resistance as well as cross-resistance to other antibiotics. Some RIF-resistant isolates have been seen to be cross-resistant to INH [OR: 0.27, 95% confidence interval (CI): 0.08109–0.9437; P=0.041] and MOX (OR: 0.8718, 95% CI: 0.7114–0.9708; P=0.014). Though resistance to RIF has been seen to be associated with INH/MOX resistance, no such relation was observed between rifampicin-susceptible (RIFS) or RIFR and other anti-TB drugs [ethambutol (EMB), pyrazinamide (PZA), streptomycin (STR) and LFX] (Table 3). In the other factors, the statistically significant data were observed among the RIFS, RIFR, and Pre-XDR cases (OR: 0.9136, 95% CI: 0.8034–0.9841; P=0.02). However, no significant data were observed in MDR cases.
Table 3
| Characteristics | Resistant | Susceptible | P value | OR | 95% CI |
|---|---|---|---|---|---|
| Gender | 0.999 | 0.9917 | 0.9276–1.072 | ||
| Male | 105 | 7 | |||
| Female | 121 | 8 | |||
| Age, years | 0.785 | 1.011 | 0.9415–1.088 | ||
| <30 | 115 | 7 | |||
| ≥30 | 111 | 8 | |||
| Treatment history | 0.791 | 1.016 | 0.9481–1.096 | ||
| New case | 120 | 7 | |||
| Retreated | 106 | 8 | |||
| INH resistant | 0.041* | 0.27 | 0.08109–0.9437 | ||
| Yes | 125 | 14 | |||
| No | 99 | 3 | |||
| EMB resistant | 0.999 | 1.01 | 0.8769–1.077 | ||
| Yes | 35 | 2 | |||
| No | 191 | 13 | |||
| PZA resistant | 0.713 | 0.9849 | 0.8739–1.047 | ||
| Yes | 46 | 3 | |||
| No | 183 | 9 | |||
| STR resistant | 0.373 | 1.069 | 0.9148–4.386 | ||
| Yes | 23 | 0 | |||
| No | 204 | 14 | |||
| LFX resistant | 0.198 | 0.9492 | 0.8009–1.021 | ||
| Yes | 31 | 3 | |||
| No | 195 | 8 | |||
| MOX resistant | 0.014* | 0.8718 | 0.7114–0.9708 | ||
| Yes | 30 | 6 | |||
| No | 195 | 9 | |||
| MDR-TB | 0.248 | 0.9580 | 0.8796–1.021 | ||
| Yes | 76 | 5 | |||
| No | 95 | 2 | |||
| Pre-XDR | 0.02* | 0.9136 | 0.8034–0.9841 | ||
| Yes | 53 | 7 | |||
| No | 175 | 6 |
A total of 1,303 cases were included in the study, consisting of 241 Mtb positive cases and 1,062 Mtb negative cases. Among the 241 positive cases, 112 were male and 129 were female. Among the 1,062 negative cases, 400 were male and 662 were female (P=0.0160, 95% CI: 1.062–1.670, OR: 1.4). *, P<0.05 is considered statistically significant. RIF, rifampicin; OR, odds ratio; CI, confidence interval; INH, isoniazid; EMB, ethambutol; PZA, pyrazinamide; STR, streptomycin; LFX, levofloxacin; MOX, moxifloxacin; MDR, multi-drug resistant; TB, tuberculosis; XDR, extreme drug resistant; Mtb, Mycobacterium tuberculosis.
INH resistance is significantly related to the resistance to the EMB (OR: 1.874, 95% CI: 1.571–2.190; P<0.001), PZA (OR: 2.090, 95% CI: 1.793–2.460; P<0.001), STR (OR: 1.680, 95% CI: 1.307–1.968; P<0.001), LFX (OR: 1.913, 95% CI: 1.622–2.222; P<0.001), and MOX (OR: 1.773, 95% CI: 1.462–2.080; P<0.001). However, no significant data was observed in isoniazid-susceptible (INHS) and isoniazid-resistant (INHR) to the RIF. While among the other factors, the statistically significant data were observed in the INHS and INHR, MDR cases (OR: 17.92, 95% CI: NA to 41.62; P<0.001), and Pre-XDR (OR: 2.047, 95% CI: 1.736–2.435; P<0.001) (Table 4).
Table 4
| Characteristics | INH-resistant | INH-susceptible | P value | OR | 95% CI |
|---|---|---|---|---|---|
| Gender | 0.43 | 0.80 | 0.7277–1.136 | ||
| Male | 61 | 51 | |||
| Female | 77 | 52 | |||
| Age, years | 0.698 | 1.11 | 0.8414–1.312 | ||
| <30 | 71 | 51 | |||
| ≥30 | 66 | 53 | |||
| Treatment history | 0.896 | 0.93 | 0.7810–1.216 | ||
| New case | 71 | 55 | |||
| Retreated | 66 | 48 | |||
| RIF-resistant | 0.103 | 0.6969 | 0.5667–1.031 | ||
| Yes | 126 | 100 | |||
| No | 12 | 3 | |||
| EMB-resistant | <0.001*** | 1.874 | 1.571–2.190 | ||
| Yes | 35 | 2 | |||
| No | 103 | 101 | |||
| PZA-resistant | <0.001*** | 2.090 | 1.793–2.460 | ||
| Yes | 48 | 1 | |||
| No | 90 | 102 | |||
| STR-resistant | <0.001*** | 1.680 | 1.307–1.968 | ||
| Yes | 20 | 2 | |||
| No | 118 | 100 | |||
| LFX-resistant | <0.001*** | 1.913 | 1.622–2.222 | ||
| Yes | 33 | 1 | |||
| No | 105 | 102 | |||
| MOX-resistant | <0.001*** | 1.773 | 1.462–2.080 | ||
| Yes | 33 | 3 | |||
| No | 106 | 99 | |||
| MDR-TB | <0.001*** | 17.92 | NA to 41.62 | ||
| Yes | 133 | 11 | |||
| No | 5 | 92 | |||
| Pre-XDR | <0.001*** | 2.047 | 1.736–2.435 | ||
| Yes | 57 | 3 | |||
| No | 84 | 97 |
***, P<0.001 is considered highly statistically significant. INH, isoniazid; OR, odds ratio; CI, confidence interval; RIF, rifampicin; EMB, ethambutol; PZA, pyrazinamide; STR, streptomycin; LFX, levofloxacin; MOX, moxifloxacin; MDR, multi-drug resistant; TB, tuberculosis; XDR, extreme drug resistant.
FQ resistance is significantly related to resistance to the INH (OR: 11.38, 95% CI: 3.312–15.66; P<0.001), EMB (OR: 2.810, 95% CI: 1.855–4.082; P<0.001), and PZA (OR: 2.939, 95% CI: 1.971–4.290; P<0.001). No significant association was seen between FQ-susceptible (FQS) or FQR and resistance to RIF and STR. The rate of FQR was seen to be associated with gender (OR, 0.53; 95% CI: NA to 0.9616; P=0.040) and age (OR, 2.143; 95% CI: NA to 2.874; P=0.012), with higher incidents of FQR in females and individuals below 30 years of age (Table 5). This difference was due to healthcare-seeking behavior, treatment adherence, or exposure to risk factors, which could potentially contribute to varying rates of drug resistance between sexes. Incidence of FQR was also seen to be higher among MDR (OR: 21.38, 95% CI: 4.576–38.93; P<0.001) and Pre-XDR (OR: 34.99, 95% CI: 15.31–81.62; P<0.001) cases.
Table 5
| Characteristics | FQ-resistant | FQ-susceptible | P value | OR | 95% CI |
|---|---|---|---|---|---|
| Gender | 0.040* | 0.53 | NA to 0.9616 | ||
| Male | 22 | 90 | |||
| Female | 41 | 88 | |||
| Age, years | 0.012* | 2.143 | NA to 2.874 | ||
| <30 | 42 | 88 | |||
| ≥30 | 20 | 91 | |||
| Treatment history | 0.305 | 0.71 | 0.5131–1.197 | ||
| New case | 29 | 96 | |||
| Retreated | 34 | 81 | |||
| RIF-resistant | 0.229 | 0.50 | 0.3667–1.312 | ||
| Yes | 57 | 169 | |||
| No | 6 | 9 | |||
| INH-resistant | <0.001*** | 11.38 | 3.312–15.66 | ||
| Yes | 57 | 81 | |||
| No | 6 | 97 | |||
| EMB-resistant | <0.001*** | 2.810 | 1.855–4.082 | ||
| Yes | 21 | 16 | |||
| No | 41 | 162 | |||
| PZA-resistant | <0.001*** | 2.939 | 1.971–4.290 | ||
| Yes | 27 | 22 | |||
| No | 36 | 156 | |||
| STR-resistant | 0.325 | 1.58 | 0.7189–2.336 | ||
| Yes | 8 | 15 | |||
| No | 55 | 163 | |||
| MDR-TB | <0.001*** | 21.38 | 4.576–38.93 | ||
| Yes | 58 | 85 | |||
| No | 3 | 94 | |||
| Pre-XDR | <0.001*** | 34.99 | 15.31–81.62 | ||
| Yes | 58 | 2 | |||
| No | 5 | 176 | |||
*, P<0.05 is considered statistically significant; ***, P<0.001 is considered highly statistically significant. FQ, fluoroquinolones; OR, odds ratio; CI, confidence interval; RIF, rifampicin; INH, isoniazid; EMB, ethambutol; PZA, pyrazinamide; STR, streptomycin; MDR, multi-drug resistant; TB, tuberculosis; XDR, extreme drug resistant.
The treatment outcomes of TB-positive patients are depicted in Figure 2. One hundred and thirty-six of the 241 TB patients who tested positive were successfully cured after treatment and 30 died. Furthermore, the therapy of the patients was reported as unsuccessful, and there were 17 cases of loss to follow-up.
Discussion
The evolution of DR-TB threatens global TB control efforts, which warrants increased surveillance and identification of drug resistance patterns to devise ways to counter drug resistance mechanisms in Mtb. This could be achieved by identifying genetic determinants of drug resistance in this pathogen and enhancing target-based development of therapeutics as well as identification of effective combination regimens.
This study was conducted at the Center for Biotechnology and Microbiology, University of Swat (Saidu Teaching Hospital) to assess the prevalence of primary MDR-TB and study the utility of LPA for its rapid detection among new TB-positive cases. This study also looks at the most common genetic mutations implicated in RIF, INH, and FQ resistance in the study population.
It was observed in this study that female patients constitute 54% of the study population in contrast to their male counterparts, suggesting that females are at higher risk of contracting TB in this study (11). This difference was due to healthcare-seeking behavior, treatment adherence, or exposure to risk factors. Which could potentially contribute to varying rates of drug resistance between sexes. The highest incidence of TB was found among individuals within 15–24 years age range. This finding corroborates a previous report (12) in which high rate of TB was reported in similar age groups, 1–20 years (28.1%) and 21–40 years (40%). New cases constitute over half of the TB-positive cases identified in this study (Table 1), stressing the public health significance of TB. However, this deviates from a previous report (13) in which the TB-positive cases were mostly related to retreated rather than new cases. We presume that this difference could have come from different sample sizes or different geographical regions.
MDR-TB makes patients prone to treatment failure or complications that come with prolonged therapy, particularly when using conventional first- and second-line regimens. High incidence of DR-TB was observed among the studied cases, including high level of MDR-TB (Figure 1). Of the 241 TB-positive cases, there were 97 cases of mono-resistance, 84 MDR cases, 57 Pre-XDR cases and 3 XDR cases. Genetic determinants of such resistance were identified, including those with multiple mutations on the same gene (Table 2).
There is evidence of resistance to RIF, INH and FQs among the studied patients which influences the possibility for treatment failure using first-line and perhaps second-line regimens. For instance, rpoB, a determinant of RIF resistance in Mtb (14) has been seen to be the principal determinant of RIF resistance within the studied population, including a mutation identified within RRDR (15). The most prevalent mutation seen to be associated with RIFR is S531L in the rpoB gene (16). The high frequency of the S531L mutation may be attributed to its low fitness cost, which affects both its strong environmental selection and transmissibility. RIFR may serve as an alternative marker for MDR-TB as the majority of RIFR Mtb isolates are resistant to INH (17). However, in regions with low prevalence of RIFR, fast testing for RIFR alone cannot reliably predict MDR-TB (18). These tests might be useful in an MDR-TB management plan in regions with high RIFR and MDR-TB prevalence.
Likewise, katG and inhA have been previously shown to mediate resistance to INH in Mtb (15,19) which has been seen to be the case with INH-resistant TB among the studied population. In this study, a mutation at the −15 nucleic acid position of the promoter region of inhA (C15T) (74 cases) is seen to be most commonly associated with INH resistance. Mutations in the promoter region of inhA, particularly the −15C→T mutation, are also the cause of high-level resistance to ethionamide (ETH) and prothionamide (PTO) in addition to low-level resistance to INH (20) and could therefore form the basis for exclusion of ETH and PTO from MDR-TB treatment regimens. Of the katG mutations implicated in INH resistance, 75–90% have been said to occur on the 315th codon of the protein, often leading to Ser315Thr and Ser315ILe mutations. However, there is a geographical variation in the prevalence of katG and inhA mutations (19). It was seen that 46% of INH-resistant isolates in this study harbored a mutation within the katG gene.
GenoType MTBDRsl assays were used to determine the prevalence of FQ resistance among the isolates in this study. A total of 63/241 (26%) were found to be resistant to FQs. FQ-resistant TB in the studied population has been seen to be due to mutations within the quinolone resistance-determining region of gyrA gene which corroborates previous reports (21,22). Hybridization pattern analysis revealed that most isolates harbour a mutation in gyrA locus with substitutions at position 94 (D94Y) being more prevalent. However, in the presence of D94G mutation, both LFX and MOX have been seen to be ineffective. D94A and D94N were additional mutations seen to be harboured by the isolates and isolates with D94A mutations could be susceptible to MOX at higher doses but resistant to LFX. However, D94N/Y confers resistance to both LFX and MOX. These findings corroborate previous reports (23-25). FQ resistance was seen to be most commonly associated with the D94G mutation in MDR and XDR cases. This mutation confers high-level FQ resistance, including resistance to the fourth-generation MOX. D94Y mutation has been seen to be prevalent among newly diagnosed TB cases with FQ resistance, though high-dose MOX remains the drug of choice in such cases. In contrast, D94G was seen to be common among treatment failure and relapse cases. The prevalence of FQ resistance is also high in other regions of Pakistan (26,27) as well as neighboring countries of India (16,28), China (17,29), and Bangladesh (18,30).
Mutations within regulatory and coding regions of drug resistance-determining genes have been implicated in resistance to other antibiotics (21,31-34). The incidence of resistance to other anti-tubercular drugs is quite evident in this study (Tables 3-5), which implies the prevalence of MDR-TB in the studied population. For instance, a significant level of co-resistance was observed between RIF and INH as well as between RIF and MOX (P<0.05), which are important FLDs and SLDs. Co-resistance to RIF and INH potentially renders the first-line drugs PZA and EMB ineffective, and resistance to MOX may reduce the effectiveness of the second-line regimens. This will increase the burden of MDR-TB cases and eventually that of Pre-XDR-TB. Significant level of co-resistance (P<0.001) was also seen between INH and five other drugs (EMB, PZA, STR, LFX and MOX) (Table 4). Co-resistance between INH and other drugs may possibly be due to the high spontaneous mutation rate of INH or due to inhA as such mutations have been previously implicated in co-resistance to INH and PTO as they have the same target, InhA (35,36). The accumulation of drug resistance mutations occurs in a cumulative manner, so the presence of co-resistant strains implies that these strains have been subject to multiple therapies over the lifetime of the strain lineage.
Of the 241 TB-positive cases in the study population, more than half were successfully cured following short-term/long-term treatment using appropriate regimens, choice of which was determined by severity of drug resistance. Short-term treatment was administered for 9–11 months using the combination MOX + ETH + clofazimine + PZA + INH + EMB, whereas long-term treatment was administered for 18 months using the combination LFX+ linezolid + clofazimine + decylocerin + PZA. However, others succumbed to TB, had failed treatment, were lost to follow up or are still under treatment perhaps due to extensive drug resistance. Treatment failed in 2% of the patients and 22% are still under treatment. The treatment failure could be the result of the limitations of LPA to assay mutations for resistance the latest group of anti-TB drugs (linezolid, delamanid, etc.) or novel mutations for resistance to FLDs and SLDs. This paints a rather stark picture of the extent of resistance in these patients and warrants controlled use of antibiotics among TB patients.
Conclusions
This study establishes the combined utility of GeneXpert and LPA hybridization methods in rapid diagnosis of DR-TB. It is unsurprising that DR-TB is prevalent in Pakistan since the country is one of those with high TB burden. This certainly warrants serious countermeasures to ensure effective prevention and control of both DS- and DR-TB. These could include repurposing of drugs, target-based development of anti-TB drugs, identification of effective, safe combination regimens as well as development of effective anti-TB vaccines. This study also provides a substantial sample size and comprehensive analysis of the mutational patterns in Mtb strains from Pakistan. It can contribute valuable data to the global understanding of MDR-TB epidemiology and genotypic characteristics. The study’s findings regarding the prevalence of MDR isolates, specific mutations in the rpoB, katG, and inhA genes, and the effectiveness of GenoType MTBDRplus and MTBDRsl assays can potentially provide important insights into the local situation and contribute to the global knowledge and fast molecular diagnosis based on MDR-TB. However, the current study had a limited number of data, and further studies are warranted in the other areas of Pakistan, to access the accurate number and burden of TB.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-138/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-138/dss
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-138/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-138/coif). The authors have no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Abbasi MA. Guidelines for the treatment of drug resistant Tuberculosis: the 2018 revision. J Ayub Med Coll Abbottabad 2018;30:493-4.
- WHO. Global Tuberculosis report. Geneva: World Health Organization; 2022.
- Hameed HMA, Fang C, Liu Z, et al. Characterization of Genetic Variants Associated with Rifampicin Resistance Level in Mycobacterium tuberculosis Clinical Isolates Collected in Guangzhou Chest Hospital, China. Infect Drug Resist 2022;15:5655-66. [Crossref] [PubMed]
- Islam MM, Tan Y, Hameed HMA, et al. Phenotypic and Genotypic Characterization of Streptomycin-Resistant Multidrug-Resistant Mycobacterium tuberculosis Clinical Isolates in Southern China. Microb Drug Resist 2020;26:766-75. [Crossref] [PubMed]
- Hameed HMA, Tan Y, Islam MM, et al. Phenotypic and genotypic characterization of levofloxacin- and moxifloxacin-resistant Mycobacterium tuberculosis clinical isolates in southern China. J Thorac Dis 2019;11:4613-25. [Crossref] [PubMed]
- Javaid MA, Ahmed AS, Durand R, et al. Saliva as a diagnostic tool for oral and systemic diseases. J Oral Biol Craniofac Res 2016;6:66-75. [Crossref] [PubMed]
- Domínguez J, Boettger EC, Cirillo D, et al. Clinical implications of molecular drug resistance testing for Mycobacterium tuberculosis: a TBNET/RESIST-TB consensus statement. Int J Tuberc Lung Dis 2016;20:24-42. [Crossref] [PubMed]
- Yacoob FL, Philomina Jose B, Karunakaran Lelitha SD, et al. Primary Multidrug Resistant Tuberculosis and Utility of Line Probe Assay for Its Detection in Smear-Positive Sputum Samples in a Tertiary Care Hospital in South India. J Pathog 2016;2016:6235618. [Crossref] [PubMed]
- Jaleta KN, Gizachew M, Gelaw B, et al. Rifampicin-resistant Mycobacterium tuberculosis among tuberculosis-presumptive cases at University of Gondar Hospital, northwest Ethiopia. Infect Drug Resist 2017;10:185-92. [Crossref] [PubMed]
- Datema TA, Oskam L, Engelberts MF, et al. Global laboratory initiative tool for a stepwise process towards tuberculosis laboratory accreditation. Int J Tuberc Lung Dis 2012;16:704-5. [Crossref] [PubMed]
- Lalokhil MS, Khan A, Adnan M, et al. Prevalence of pulmonary tuberculosis in district Mardan Khyber Pakhtunkhwa, Pakistan. Microbiol Curr Res 2019;3:1-7.
- Uddin S, Khan FU, Ahmad T, et al. Two Years of Retrospective Study on the Incidence of Tuberculosis in Dir Lower Valley, Khyber Pakhtunkhwa, Pakistan. Hosp Pharm 2018;53:344-9. [Crossref] [PubMed]
- Ali S, Khan MT, Khan AS, et al. Prevalence of Multi-Drug Resistant Mycobacterium tuberculosis in Khyber Pakhtunkhwa - A High Tuberculosis Endemic Area of Pakistan. Pol J Microbiol 2020;69:1-5. [Crossref] [PubMed]
- Telenti A, Imboden P, Marchesi F, et al. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet 1993;341:647-50. [Crossref] [PubMed]
- Ramaswamy SV, Reich R, Dou SJ, et al. Single nucleotide polymorphisms in genes associated with isoniazid resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 2003;47:1241-50. [Crossref] [PubMed]
- Sharma AK, Gupta N, Kala DK, et al. A study on pattern of resistance to second line anti tubercular drugs among multi drug resistant tuberculosis patients. Indian J Tuberc 2018;65:233-6. [Crossref] [PubMed]
- Wang Z, Xie T, Mu C, et al. Performance of Sequencing in Predicting Ofloxacin Resistance in Mycobacterium tuberculosis from Positive Bactec MGIT 960 Cultures. Ann Clin Lab Sci 2018;48:69-74.
- Rigouts L, Coeck N, Gumusboga M, et al. Specific gyrA gene mutations predict poor treatment outcome in MDR-TB. J Antimicrob Chemother 2016;71:314-23. [Crossref] [PubMed]
- Tessema B, Beer J, Emmrich F, et al. Analysis of gene mutations associated with isoniazid, rifampicin and ethambutol resistance among Mycobacterium tuberculosis isolates from Ethiopia. BMC Infect Dis 2012;12:37. [Crossref] [PubMed]
- Havlicek J, Dachsel B, Slickers P, et al. Rapid Microarray-Based Detection of Rifampin, Isoniazid, and Fluoroquinolone Resistance in Mycobacterium tuberculosis by Use of a Single Cartridge. J Clin Microbiol 2018;56:e01249-17. [Crossref] [PubMed]
- Nosova EY, Bukatina AA, Isaeva YD, et al. Analysis of mutations in the gyrA and gyrB genes and their association with the resistance of Mycobacterium tuberculosis to levofloxacin, moxifloxacin and gatifloxacin. J Med Microbiol 2013;62:108-13. [Crossref] [PubMed]
- Pantel A, Petrella S, Veziris N, et al. Extending the definition of the GyrB quinolone resistance-determining region in Mycobacterium tuberculosis DNA gyrase for assessing fluoroquinolone resistance in M. tuberculosis. Antimicrob Agents Chemother 2012;56:1990-6. [Crossref] [PubMed]
- Avalos E, Catanzaro D, Catanzaro A, et al. Frequency and geographic distribution of gyrA and gyrB mutations associated with fluoroquinolone resistance in clinical Mycobacterium tuberculosis isolates: a systematic review. PLoS One 2015;10:e0120470. [Crossref] [PubMed]
- Singhal R, Reynolds PR, Marola JL, et al. Sequence Analysis of Fluoroquinolone Resistance-Associated Genes gyrA and gyrB in Clinical Mycobacterium tuberculosis Isolates from Patients Suspected of Having Multidrug-Resistant Tuberculosis in New Delhi, India. J Clin Microbiol 2016;54:2298-305. [Crossref] [PubMed]
- Von Groll A, Martin A, Jureen P, et al. Fluoroquinolone resistance in Mycobacterium tuberculosis and mutations in gyrA and gyrB. Antimicrob Agents Chemother 2009;53:4498-500. [Crossref] [PubMed]
- Akhtar AM, Arif MA, Kanwal S, et al. Prevalence and drug resistance pattern of MDR TB in retreatment cases of Punjab, Pakistan. J Pak Med Assoc 2016;66:989-93.
- Jabeen K, Shakoor S, Malik F, et al. Fluoroquinolone resistance in Mycobacterium tuberculosis isolates from Pakistan 2010-2014: Implications for disease control. Int J Mycobacteriol 2015;4:47-8. [Crossref] [PubMed]
- Desai U, Joshi JM. Extrapulmonary drug-resistant tuberculosis at a drug-resistant tuberculosis center, Mumbai: Our experience - Hope in the midst of despair! Lung India 2019;36:3-7. [Crossref] [PubMed]
- Gao Y, Zhang Z, Deng J, et al. Multi-center evaluation of GenoType MTBDRsl line probe assay for rapid detection of pre-XDR and XDR Mycobacterium tuberculosis in China. J Infect 2018;77:328-34. [Crossref] [PubMed]
- Kamal SM, Hossain A, Sultana S, et al. Anti-tuberculosis drug resistance in Bangladesh: reflections from the first nationwide survey. Int J Tuberc Lung Dis 2015;19:151-6. [Crossref] [PubMed]
- Machado D, Perdigão J, Ramos J, et al. High-level resistance to isoniazid and ethionamide in multidrug-resistant Mycobacterium tuberculosis of the Lisboa family is associated with inhA double mutations. J Antimicrob Chemother 2013;68:1728-32. [Crossref] [PubMed]
- Mboowa G, Namaganda C, Ssengooba W. Rifampicin resistance mutations in the 81 bp RRDR of rpoB gene in Mycobacterium tuberculosis clinical isolates using Xpert®MTB/RIF in Kampala, Uganda: a retrospective study. BMC Infect Dis 2014;14:481. [Crossref] [PubMed]
- Sowajassatakul A, Prammananan T, Chaiprasert A, et al. Molecular characterization of amikacin, kanamycin and capreomycin resistance in M/XDR-TB strains isolated in Thailand. BMC Microbiol 2014;14:165. [Crossref] [PubMed]
- Thirumurugan R, Kathirvel M, Vallayyachari K, et al. Molecular analysis of rpoB gene mutations in rifampicin resistant Mycobacterium tuberculosis isolates by multiple allele specific polymerase chain reaction in Puducherry, South India. J Infect Public Health 2015;8:619-25. [Crossref] [PubMed]
- Islam MM, Tan Y, Hameed HMA, et al. Detection of novel mutations associated with independent resistance and cross-resistance to isoniazid and prothionamide in Mycobacterium tuberculosis clinical isolates. Clin Microbiol Infect 2019;25:1041.e1-7. [Crossref] [PubMed]
- Khan Z, Ualiyeva D, Jamal K, et al. Molecular diagnostics and potential therapeutic options for Mycobacterium tuberculosis: Where we stand. Medicine in Omics 2023;8:100022.



