
Identification of potential biomarkers associated with CD4+ T cell infiltration in myocardial ischemia-reperfusion injury using bioinformation analysis
Highlight box
Key findings
• CLEC5A is a hub gene associated with CD4+ T cells in myocardial ischemia-reperfusion injury.
What is known and what is new?
• Immune cell infiltration plays an important role in myocardial ischemia-reperfusion injury. Infiltration of CD4+ T cells can lead to increased myocardial damage.
• Our analysis identified a potential association between CLEC5A expression and CD4+ T cell infiltration in myocardial ischemia-reperfusion injury.
What is the implication, and what should change now?
• CLEC5A is closely associated with CD4+ T cell recruitment in ischemia-reperfusion injury. These findings will shed light on the clarification of the immunological mechanism of ischemia-reperfusion injury and provide new targets for subsequent clinical treatment.
Introduction
At present, there is no effective treatment for myocardial ischemia-reperfusion injury (MIRI), which exacerbates energy metabolism disorder, myocardial structure damage, and functional injury (1,2). After coronary recanalization in the infarcted area, arrhythmia, decline of cardiac function, and sudden cardiac death often occur clinically. The pathogenesis of MIRI is complex, involving oxidative damage (3,4), inflammatory response (5,6), calcium overload (7,8), and other processes. Many mechanisms of MIRI are unclear, especially the role of the immune microenvironment in MIRI remains to be further explored. Previous studies have shown that CD4+ T cells are involved in this pathophysiological process (9,10). Further study on the mechanism of T cells can provide new ideas for the treatment of ischemia-reperfusion injury (IRI).
Adaptive immune responses, particularly those involving CD4+ T cells, play an important role in ischemia-reperfusion. Several studies have used CD4+ T cell-deficient mice and demonstrated a critical role for CD4+ T cells in IRI and infarct healing (11,12). However, there is a lack of suitable biomarkers for predicting the extent of CD4+ T cell infiltration in ischemia-reperfusion. The use of biosignature analysis to solve challenging biological problems will not only help advance basic research, but may also provide new breakthroughs in biomedical applications.
C-type lectins are the largest and most diverse family of mammalian carbohydrate-binding proteins. In previous studies, they have been shown to interact with various endogenous and exogenous ligands and participate in important physiological processes such as immune defense, immune stability, and immune monitoring, so as to maintain bodily homeostasis (13,14). CLEC5A, a type II transmembrane protein belonging to the type C lectin family, can recognize endogenous ligands, such as self-carbohydrates, proteins, or lipids, to control homeostasis and tissue damage. CLEC5A is associated with the progression of a variety of acute and chronic inflammatory diseases. However, the detailed mechanism of CLEC5A and CD4+ T cell infiltration in MIRI has not been determined.
In this study, we assessed the immune microenvironment of each sample. Based on the increase in CD4+ T-cell infiltration after 6 hours of reperfusion, a series of bioinformatics analyses were performed using the available data, providing a reliable basis for exploring the molecular mechanisms of CD4+ T-cell infiltration in MIRI. We present this article in accordance with the STREGA reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1335/rc).
Methods
Data collection
In this study, we compared the gene expression profiles of the myocardial tissue of mice treated with reperfusion after ischemia at 0, 6, 24, and 72 hours of ischemia/reperfusion (I/R) from the Gene Expression Omnibus (GEO; n=4 per group) and downloaded and collected high-throughput gene expression dataset (No. GSE160516). The gene probe was transformed into gene symbols according to the annotation information for GPL23038 [Clariom_S_Mouse] Affymetrix Clariom S Assay (Affymetrix, Santa Clara, CA, USA), Mouse (includes Pico assay) and normalized for differentiation analysis.
Analysis of infiltrating immune cells
ImmuCellAI-mouse (Immune Cell Abundance Identifier for mouse) is a tool to estimate the abundance of 36 immune cells based on gene expression profile from RNA sequencing (RNA-Seq) or microarray data (15). To ensure the prediction accuracy, the abundance of cells in 3 layers was predicted separately by calculating the single-sample gene set enrichment analysis (ssGSEA) enrichment score of the expression deviation profile per cell type. Besides, ImmuCellAI-mouse can be applied to estimate the difference of immune cell infiltration in diverse groups.
Co-expression network construction by WGCNA
We used WGCNA to define the specific gene modules representing each experimental group and the CD4+ T cell compositions. First, samples were clustered to assess the presence of any obvious outliers. Second, the automatic network construction function was used to construct the co-expression network. To make the co-expression network align with the distribution of scale-free network, a soft-thresholding power was computed with the pickSoftThreshold function. Third, hierarchical clustering and the dynamic tree cut function were employed to identify modules. Subsequently, a dynamic tree-cutting algorithm was utilized to establish module assignments. Module eigengenes (MEs) were computed using the MEs function. Pearson correlation coefficients between ME and each experimental group and the CD4+ T cell compositions were evaluated. After co-expression network construction by WGCNA, MEturquoise genes were selected for further analysis.
Differential expression analyses of CD4+ T cell-related genes
By Wilcoxon test in the R language (R Foundation for statistical Computing, Vienna, Austria), the differentially expressed genes (DEGs) between the IR6h and IR24h groups were identified. The cut-off criteria for the differential gene are adjusted P value (adj. P) <0.01 and false discovery rate (FDR) <1. We acquired 578 CD4+ T cell-related genes differential expression between IR6h and IR24h group. The intersection of the CD4 T cell-related DEGs and MEturquoise genes, as visualized in a Venn diagram, were selected for further analysis.
Construction of a protein-protein interaction (PPI) network
We used the Metascape database (https://metascape.org) to establish a PPI network to visualize the connections between the target genes. Metascape provides a rather unique PPI network analysis capability. To infer more biologically interpretable results, Metascape applies a mature complex identification algorithm called Molecular Complex Detection (MCODE) to automatically extract protein complexes embedded in such large network. The min overlap =3, P value cutoff =0.01, min enrichment =1.5 were regarded as standard.
Functional enrichment analysis
To identify pathways significantly enriched between IR6h and IR24h, we performed Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses. Meanwhile, we performed ssGSEA to derive the enrichment score of each immune-related term according to the “clusterprofiler” package. P value <0.05 was considered statistically significant.
Statistical analysis
The data was processed using the PERL programming language (Version 5.32.0; http://www.perl.org). All statistical analyses were performed using the R software (version 4.1.2; https://www.r-project.org/). P value <0.05 was considered to indicate statistical significance, if not otherwise stated.
Results
CD4+ T cell-associated module identification by WGCNA
Previous studies have found that CD4+ T cells are involved in reperfusion injury after myocardial ischemia (9,10). The changes of CD4+ T cells infiltration with the time of reperfusion are shown in Figure 1A. We noticed that CD8+ T cells did not play a significant role in this process (Figure 1B). At the same time, at the transcriptome level, caspase family members also showed differences at different time points after reperfusion (Figure 1C). The immune infiltrating abundance of 36 immune cells were predicted using ImmuCellAI-mouse. In the R package WGCNA, we constructed co-expression networks with reperfusion time and CD4+ T cells infiltration score (Figure 1D-1I). CD4+ T cells, which presented statistical significance between IR6h and IR24h, were used to construct a co-expression network with 12 as the soft thresholding power β. The changes of gene expression and T cell infiltration in each module with reperfusion time were observed and compared, and the MEturquoise module were selected for further analysis.
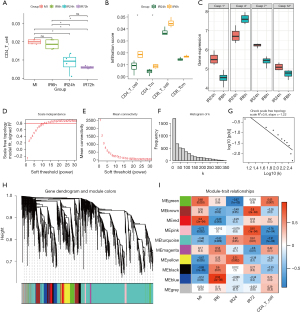
Identification of CD4+ T cell-related genes and construction of a PPI network
By Wilcoxon test in the R language, the 578 DEGs between IR6h and IR24h samples were identified (Figure 2A,2B). The 406 intersecting genes of DEGs and MEturquoise module genes were shown by Venn diagram (Figure 2C). Then, the PPI network was constructed by using these intersecting genes, because these DEGs have been shown to relate to the interacting proteins (Figure 2D). The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) furnishes original reliable protein data for consequent analysis. Through the PPI network, a total of 12 hub genes were identified as candidate genes for further analysis.
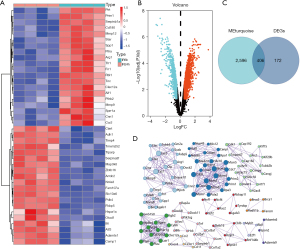
Functional enrichment analysis of intersecting genes
To further comprehend the latent functions of the 406 intersecting genes in MIRI, we performed GO, gene set variation analysis (GSVA), gene set enrichment analysis (GSEA), and KEGG enrichment analyses. KEGG pathway analysis demonstrated that the intersection genes were mainly involved in chemokine signaling pathway and cell cycle (Figure 3A,3B). In the results of GSVA and GSEA analysis, hypoxia and immune checkpoints were in a conspicuous position (Figure 3C,3D). The top 10 components of biological process (BP), cellular component (CC), and molecular function (MF) are illustrated in Figure 3E. Interestingly, we discovered that the cell activation involved in immune response and reactive oxygen species metabolic process also appeared in the results.
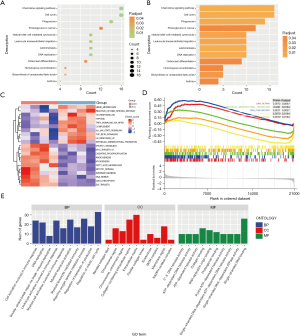
CLEC5A is closely related to CD4+ T cell infiltration after MIRI
Through the correlation analysis between the 12 hub genes found in the PPI network and CD4+ T cell infiltration fraction, we identified the core gene CLEC5A (Figure 4A). Moreover, the change of CLEC5 expression after reperfusion was shown to have a very high correlation with the degree of T cell infiltration (Figure 4B). Since we found the important role of immune checkpoints through enrichment analysis, we paid more attention to the role of CLEC5A in immunity. According to the expression of CLEC5A, we divided all samples into two groups and analyzed the infiltration of all immune cells (Figure 4C). In addition to CD4+ T cells, CLEC5A is also associated with macrophages and neutrophils. At the same time, we found that CLEC5A was also associated with a variety of chemokines, which reasonably explained the correlation between CLEC5A and immune infiltration (Figure 4D).
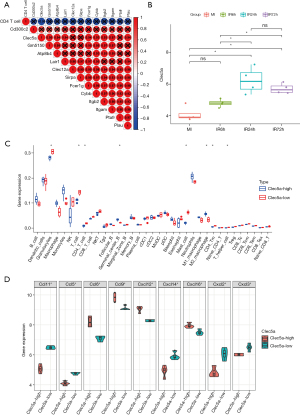
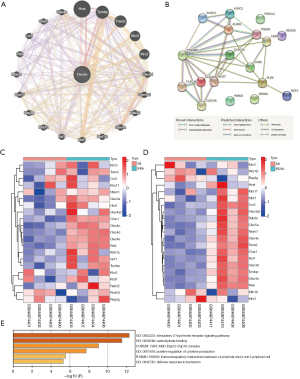
Functional analysis of CLEC5A and its related genes
GeneMANIA was used to identify the top 20 CLEC5A-related genes of the hub gene and construct a gene interaction network (Figure 5A). At the same time, the PPI network was constructed (Figure 5B). We then observed the expression of these genes at different times after ischemia. We used IR6h and IR24h to analyze the difference with the MIRI group (Figure 5C,5D). There were significant differences in some genes after reperfusion. Then, the 20 genes were analyzed by functional enrichment using the GeneMANIA website. The results showed that it was mainly related to C-type lectin (Figure 5E).
Discussion
IRI is often a component of clinical events such as cardiac arrest, resuscitation, and reperfusion after coronary artery occlusion (16). In clinical practice, it has been found that restoring blood perfusion may cause more serious damage than ischemia, and this phenomenon exists in different species (17). This study shows that CD4+ T cells are involved in MIRI in mice, but not in the early MI/R (MI/R for 6 h, IR6h). In order to screen the genes most closely related to T cell immunity, we further combined with the immune infiltration of mice, and screened CLEC5A as the key target gene through WGCNA and gene expression difference analysis. Previous studies on CLEC5A have mostly been related to the immune response caused by microbial virus or tumor, but research on the immune effect of MIRI has been neglected (18,19).
The pathophysiological mechanisms of myocardial ischemia-reperfusion injury are complex, but progress is being made in this area of research to explore more effective preventive and therapeutic approaches to achieve the goal of mitigating the impact of myocardial reperfusion injury on heart health. Existing research findings are mainly related to inflammatory responses, oxidative stress, and calcium overload (20-22). Researchers are also currently exploring new approaches such as the use of antioxidants, inhibition of inflammatory responses, stem cell therapy or gene therapy to protect cardiomyocytes (23-25). This will hopefully improve the quality of life and prognosis of heart disease patients.
Adaptive immune responses, especially those involving CD4+ T cells, are important for wound healing. In recent years, many robust evidences have been yielded from animal models that CD4+ T cells play an important role in ischemia-reperfusion (26,27). CD4+ T cells are widely involved in reperfusion injury, not limited to myocardial tissue (28). CD4+ T cells mediate hepatic neutrophil recruitment and liver injury during hepatic ischemia-reperfusion (29). Previous studies have suggested that CD4+ lymphocytes are rapidly recruited to the injured site after ischemia-reperfusion, and promote the subsequent neutrophil recruitment through an interleukin-17 (IL-17) dependent mechanism (30,31). Similarly, CD4+ T cells also play a role in renal and intestinal IRI through their immune characteristics (32,33).
CLEC5A, a type II transmembrane protein belonging to the type C lectin family, can recognize endogenous ligands, such as self-carbohydrates, proteins, or lipids, to control homeostasis and tissue damage (34). CLEC5A is associated with the progression of a variety of acute and chronic inflammatory diseases, including dengue fever (35,36), fatal shock (37,38), Crohn’s disease (39), and autoimmune arthritis (40). In addition, some studies have shown that CLEC5A may act as a biomarker in the field of cancer (41,42). A previous study found that CLEC5A knockdown prevents myocardial infarction (MI)-induced cardiac insufficiency by regulating macrophage polarization, NLRP3 inflammation, and cell death in the left ventricle of mice (43). It has been reported that lectin plays an important role in local and remote tissue injury related to gastrointestinal ischemia-reperfusion (GI/R), and expounded the correlation between lectin and complement pathway (44). Similarly, macrophage-inducible C-type lectin has been shown to induce persistent aseptic inflammation after acute renal IRI in mice, aggravating tissue injury and subsequent renal atrophy (45). However, the detailed mechanism of CLEC5A and CD4+ T cell infiltration needs to be further studied.
Admittedly, there were limitations to this study. Above all, our study included a small sample size. Most importantly, we cannot completely deny the role of other genes in recruiting or inhibiting CD4+ T cells (46). In addition, large differences were observed between the values of individual tissue samples. Secondly, for the mouse models in the data set, many experimental details were not provided in detail, and there was a lack of evaluation of the mouse MIRI model, such as the degree of myocardial injury, infarct size, ejection function, and so on. However, this does not affect the reliability of our results and conclusions, as well as the method of data analysis. Thirdly, there were relatively few studies on C-type lectins and CD4+ T cells, strong experimental evidence is lacking, and this area requires further research.
Conclusions
This study involved a bioinformatics analysis of RNA-Seq data at different time points after reperfusion. Then, a series of bioinformatics methods including PPI network and WGCNA module were used to find the hub gene related to CD4+ T cells. The correlation between C-type lectin and IRI was found by enrichment analysis. These findings will shed light on the clarification of the immunological mechanism of IRI and provide new targets for subsequent clinical treatment.
Acknowledgments
We acknowledge the contributions of all doctors from the Department of Thoracic Surgery, Affiliated Hospital of Nantong University to the study. They also gave written permission for the publication of data and conclusions.
Funding: This study was supported by grants from
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1335/rc
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1335/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1335/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Toldo S, Mauro AG, Cutter Z, et al. Inflammasome, pyroptosis, and cytokines in myocardial ischemia-reperfusion injury. Am J Physiol Heart Circ Physiol 2018;315:H1553-68. [Crossref] [PubMed]
- Cadenas S. ROS and redox signaling in myocardial ischemia-reperfusion injury and cardioprotection. Free Radic Biol Med 2018;117:76-89. [Crossref] [PubMed]
- Zhang D, Zheng N, Fu X, et al. Dl-3-n-butylphthalide attenuates myocardial ischemia reperfusion injury by suppressing oxidative stress and regulating cardiac mitophagy via the PINK1/Parkin pathway in rats. J Thorac Dis 2022;14:1651-62. [Crossref] [PubMed]
- Zhang Z, Zhou M, Liu H, et al. Protective effects of Shen Yuan Dan on myocardial ischemia-reperfusion injury via the regulation of mitochondrial quality control. Cardiovasc Diagn Ther 2023;13:395-407. [Crossref] [PubMed]
- Mauler M, Herr N, Schoenichen C, et al. Platelet Serotonin Aggravates Myocardial Ischemia/Reperfusion Injury via Neutrophil Degranulation. Circulation 2019;139:918-31. [Crossref] [PubMed]
- Sun X, Wei Z, Li Y, et al. Renal denervation restrains the inflammatory response in myocardial ischemia-reperfusion injury. Basic Res Cardiol 2020;115:15. [Crossref] [PubMed]
- Wang R, Wang M, Zhou J, et al. Calenduloside E suppresses calcium overload by promoting the interaction between L-type calcium channels and Bcl2-associated athanogene 3 to alleviate myocardial ischemia/reperfusion injury. J Adv Res 2021;34:173-86. [Crossref] [PubMed]
- Li S, Chen J, Liu M, et al. Protective effect of HINT2 on mitochondrial function via repressing MCU complex activation attenuates cardiac microvascular ischemia-reperfusion injury. Basic Res Cardiol 2021;116:65. [Crossref] [PubMed]
- Hofmann U, Frantz S. Role of T-cells in myocardial infarction. Eur Heart J 2016;37:873-9. [Crossref] [PubMed]
- Yang Z, Day YJ, Toufektsian MC, et al. Myocardial infarct-sparing effect of adenosine A2A receptor activation is due to its action on CD4+ T lymphocytes. Circulation 2006;114:2056-64. [Crossref] [PubMed]
- Mathes D, Weirather J, Nordbeck P, et al. CD4(+) Foxp3(+) T-cells contribute to myocardial ischemia-reperfusion injury. J Mol Cell Cardiol 2016;101:99-105. [Crossref] [PubMed]
- Borg N, Alter C, Görldt N, et al. CD73 on T Cells Orchestrates Cardiac Wound Healing After Myocardial Infarction by Purinergic Metabolic Reprogramming. Circulation 2017;136:297-313. [Crossref] [PubMed]
- Brown GD, Willment JA, Whitehead L. C-type lectins in immunity and homeostasis. Nat Rev Immunol 2018;18:374-89. [Crossref] [PubMed]
- Geijtenbeek TB, Gringhuis SI. Signalling through C-type lectin receptors: shaping immune responses. Nat Rev Immunol 2009;9:465-79. [Crossref] [PubMed]
- Miao YR, Xia M, Luo M, et al. ImmuCellAI-mouse: a tool for comprehensive prediction of mouse immune cell abundance and immune microenvironment depiction. Bioinformatics 2022;38:785-91. [Crossref] [PubMed]
- Nah J, Zhai P, Huang CY, et al. Upregulation of Rubicon promotes autosis during myocardial ischemia/reperfusion injury. J Clin Invest 2020;130:2978-91. [Crossref] [PubMed]
- Hausenloy DJ, Yellon DM. Myocardial ischemia-reperfusion injury: a neglected therapeutic target. J Clin Invest 2013;123:92-100. [Crossref] [PubMed]
- Wang Q, Shi M, Sun S, et al. CLEC5A promotes the proliferation of gastric cancer cells by activating the PI3K/AKT/mTOR pathway. Biochem Biophys Res Commun 2020;524:656-62. [Crossref] [PubMed]
- Cheng YL, Lin YS, Chen CL, et al. Activation of Nrf2 by the dengue virus causes an increase in CLEC5A, which enhances TNF-α production by mononuclear phagocytes. Sci Rep 2016;6:32000. [Crossref] [PubMed]
- Pagliaro P, Penna C. Inhibitors of NLRP3 Inflammasome in Ischemic Heart Disease: Focus on Functional and Redox Aspects. Antioxidants (Basel) 2023;12:1396. [Crossref] [PubMed]
- Lambert JP, Luongo TS, Tomar D, et al. MCUB Regulates the Molecular Composition of the Mitochondrial Calcium Uniporter Channel to Limit Mitochondrial Calcium Overload During Stress. Circulation 2019;140:1720-33. [Crossref] [PubMed]
- Okada H, Lai NC, Kawaraguchi Y, et al. Integrins protect cardiomyocytes from ischemia/reperfusion injury. J Clin Invest 2013;123:4294-308. [Crossref] [PubMed]
- Brown DA, Sabbah HN, Shaikh SR. Mitochondrial inner membrane lipids and proteins as targets for decreasing cardiac ischemia/reperfusion injury. Pharmacol Ther 2013;140:258-66. [Crossref] [PubMed]
- Li X, Ou W, Xie M, et al. Nanomedicine-Based Therapeutics for Myocardial Ischemic/Reperfusion Injury. Adv Healthc Mater 2023;12:e2300161. [Crossref] [PubMed]
- Vagnozzi RJ, Maillet M, Sargent MA, et al. An acute immune response underlies the benefit of cardiac stem cell therapy. Nature 2020;577:405-9. [Crossref] [PubMed]
- Khanh Vu TH, Chen H, Pan L, et al. CD4(+) T-Cell Responses Mediate Progressive Neurodegeneration in Experimental Ischemic Retinopathy. Am J Pathol 2020;190:1723-34. [Crossref] [PubMed]
- Uehara M, Solhjou Z, Banouni N, et al. Ischemia augments alloimmune injury through IL-6-driven CD4(+) alloreactivity. Sci Rep 2018;8:2461. [Crossref] [PubMed]
- Zhang B, Yin X, Lang Y, et al. Role of cellular prion protein in splenic CD4(+) T cell differentiation in cerebral ischaemic/reperfusion. Ann Clin Transl Neurol 2021;8:2040-51. [Crossref] [PubMed]
- Kuboki S, Sakai N, Tschöp J, et al. Distinct contributions of CD4+ T cell subsets in hepatic ischemia/reperfusion injury. Am J Physiol Gastrointest Liver Physiol 2009;296:G1054-9. [Crossref] [PubMed]
- Caldwell CC, Okaya T, Martignoni A, et al. Divergent functions of CD4+ T lymphocytes in acute liver inflammation and injury after ischemia-reperfusion. Am J Physiol Gastrointest Liver Physiol 2005;289:G969-76. [Crossref] [PubMed]
- Ren Y, Wang R, Jiang L, et al. Regulatory Effects of Toll-like Receptor 4 Knockout on CD4(+) and CD8(+) T Lymphocytes and Interleukin-17 During Myocardial Ischemia. Ann Clin Lab Sci 2020;50:761-8.
- Wang J, Xia S, Ren H, et al. The role and function of CD4+ T cells in hepatic ischemia-reperfusion injury. Expert Rev Gastroenterol Hepatol 2022;16:5-11. [Crossref] [PubMed]
- Yang DJ, Zhu ZX, Fu HB, et al. Hypertonic saline activates CD4+ and CD8+ T-lymphocytes in the small intestine to alleviate intestinal ischemia-reperfusion injury. Eur Rev Med Pharmacol Sci 2014;18:3069-75.
- Sung PS, Hsieh SL. CLEC2 and CLEC5A: Pathogenic Host Factors in Acute Viral Infections. Front Immunol 2019;10:2867. [Crossref] [PubMed]
- Huang YL, Chen ST, Liu RS, et al. CLEC5A is critical for dengue virus-induced osteoclast activation and bone homeostasis. J Mol Med (Berl) 2016;94:1025-37. [Crossref] [PubMed]
- Chen ST, Lin YL, Huang MT, et al. CLEC5A is critical for dengue-virus-induced lethal disease. Nature 2008;453:672-6. [Crossref] [PubMed]
- Chen ST, Li FJ, Hsu TY, et al. CLEC5A is a critical receptor in innate immunity against Listeria infection. Nat Commun 2017;8:299. [Crossref] [PubMed]
- Chen ST, Liu RS, Wu MF, et al. CLEC5A regulates Japanese encephalitis virus-induced neuroinflammation and lethality. PLoS Pathog 2012;8:e1002655. [Crossref] [PubMed]
- Elleisy N, Rohde S, Huth A, et al. Genetic association analysis of CLEC5A and CLEC7A gene single-nucleotide polymorphisms and Crohn's disease. World J Gastroenterol 2020;26:2194-202. [Crossref] [PubMed]
- Joyce-Shaikh B, Bigler ME, Chao CC, et al. Myeloid DAP12-associating lectin (MDL)-1 regulates synovial inflammation and bone erosion associated with autoimmune arthritis. J Exp Med 2010;207:579-89. [Crossref] [PubMed]
- Tong L, Li J, Choi J, et al. CLEC5A expressed on myeloid cells as a M2 biomarker relates to immunosuppression and decreased survival in patients with glioma. Cancer Gene Ther 2020;27:669-79. [Crossref] [PubMed]
- Wang R, Du X, Zhi Y. Screening of Critical Genes Involved in Metastasis and Prognosis of High-Grade Serous Ovarian Cancer by Gene Expression Profile Data. J Comput Biol 2020;27:1104-14. [Crossref] [PubMed]
- Wang X, Hu Y, Wang Y, et al. CLEC5A knockdown protects against cardiac dysfunction after myocardial infarction by suppressing macrophage polarization, NLRP3 inflammasome activation, and pyroptosis. Biochem Cell Biol 2021;99:655-65. [Crossref] [PubMed]
- Hart ML, Ceonzo KA, Shaffer LA, et al. Gastrointestinal ischemia-reperfusion injury is lectin complement pathway dependent without involving C1q. J Immunol 2005;174:6373-80. [Crossref] [PubMed]
- Lee SA, Noel S, Kurzhagen JT, et al. CD4(+) T Cell-Derived NGAL Modifies the Outcome of Ischemic Acute Kidney Injury. J Immunol 2020;204:586-95. [Crossref] [PubMed]
- Sharma AK, LaPar DJ, Zhao Y, et al. Natural killer T cell-derived IL-17 mediates lung ischemia-reperfusion injury. Am J Respir Crit Care Med 2011;183:1539-49. [Crossref] [PubMed]


