
A signature of five 7-methylguanosine-related genes is a prognostic marker for lung squamous cell carcinoma
Highlight box
Key findings
• We developed a novel N7-methylguanosine-related gene signature that can be of great utility to predict the prognosis for patients with lung squamous cell carcinoma (LSCC).
What is known and what is new?
• LSCC treatment depends on many factors including the tumor stage, resectability, performance status, and genomic alterations; however, there was not the successfully biomarkers to predict treatment outcomes and prognoses.
• In this study, we identified as an independent prognostic marker for overall survival in LSCC patients.
What is the implication, and what should change now?
• The data from the current study demonstrated that the risk model which we developed in this study could be useful for predicting the prognosis of LSCC patients. We should further promote and validate it.
Introduction
Lung cancer is one of the most commonly diagnosed cancer and the leading cause of cancer-related deaths in the world, and it continues to be a significant health burden globally (1). Histologically, up to 85% of all lung cancer cases are diagnosed as non-small cell lung cancer (NSCLC), which can be further classified as lung adenocarcinoma (LUAD) or lung squamous cell carcinoma (LSCC) (2). Etiologically, tobacco smoke and various agents that can induce DNA damage and DNA repair deficiency, are the leading risk factors for lung cancer development (3). Tobacco smoke contains thousands of constituents, many of which can damage the genomic DNA and induce normal cell transformation and carcinogenesis (4,5). Clinically, LSCC occurs more frequently in males than in females and is closely associated with tobacco smoke (6). In addition, it is characterized by distinct gene alterations and clinical outcomes compared with those of LUAD (3,7). Advanced LSCC is associated with a very poor patient prognosis due to the lack of early detection biomarkers and treatment options (8,9). Thus, further investigation of NSCLC, including LSCC, could help to elucidate the underlying molecular mechanisms in order to discover novel biomarkers. Especially early detection and prediction of prognosis and treatment outcomes as well as novel strategies for the treatment of this very deadly disease could thus be developed.
The literature indicates, epigenetic alterations, such as RNA methylation, contribute to lung cancer development and progression (10-12). N7-methylguanosine (m7G) is an endogenous methylated nucleoside found in different RNA molecules; for example, when m7G occurs in RNA messenger (mRNA), it can regulate mRNA export, translation, and splicing (13), and when m7G occurs in RNA transfer (tRNA), it can change tRNA functions to affect mRNA translation and cell growth (14). As is well known, tRNA belongs to a class of noncoding RNAs and serves as a physical link between the amino acids and the ribosomes according to the matched codon in the mRNA molecules (15,16). To date, approximately 90 different modifications that occur in tRNA molecules have been reported (17,18). Although their complete functional implications remain to be determined, m7G is one of the most frequently found in tRNA. If it occurs at position 46 of tRNA, m7G will form a tertiary base pair with C13-G22 to stabilize the 3-dimensional (3D) core of the tRNA (19-21). In addition, m7G modification of tRNA is mediated by the METTL1-WDR4 complex (22), of which METTL1 is a writer of m7G in mRNA and various noncoding RNAs, such as tRNA (23). Gene mutations or altered functions of the enzyme can contribute to human disease; for example, a mutation in the WDR4 gene will impair tRNA m7G46 methylation and cause microcephalic primordial dwarfism (24,25). Recent studies also have shown that METTL1-mediated m7G editing is important in inhibition of lung cancer cell migration (26,27). Thus, in this study, we assessed the expression of m7G-related genes as a gene signature, which were selected in base of the regulation role of these genes in non-small cell lung cancer (28-30), to predict the prognosis of LSCC patients. We present this article in accordance with the TRIPOD reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1504/rc).
Methods
Database searching and data downloading
In this study, we first searched the Cancer Genome Atlas (TCGA) database up to November 15, 2022 (https://portal.gdc.cancer.gov) to identify the differentially expressed genes (DEGs) in LSCC samples and downloaded the data of 502 LSCC patients and 49 normal adjacent lung tissues. We then searched the m7G-related genes and the complete clinicopathological information of the patients, including gender, age, tumor-node-metastasis (TNM) stage, tobacco smoking history, and survival data. We then included 29 m7G methylation-related genes to construct a risk model for LSCC (Table 1) after thoroughly searching the published literature (13) and the Gene Set Enrichment Analysis (GSEA) database (http://www.gsea-msigdb.org/gsea/login.jsp). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Table 1
| Gene | Full name |
|---|---|
| AGO2 | Argonaute RISC catalytic component 2 |
| CYFIP1 | Cytoplasmic FMR1 interacting protein 1 |
| DCP2 | Decapping mRNA 2 |
| DCPS | Decapping enzyme, scavenger |
| EIF3D | Eukaryotic translation initiation factor 3 subunit D |
| EIF4A1 | Eukaryotic translation initiation factor 4A1 |
| EIF4E | Eukaryotic translation initiation factor 4E |
| EIF4E1B | Eukaryotic translation initiation factor 4E family member 1B |
| EIF4E2 | Eukaryotic translation initiation factor 4E family member 2 |
| EIF4E3 | Eukaryotic translation initiation factor 4E family member 3 |
| EIF4G3 | Eukaryotic translation initiation factor 4 gamma 3 |
| GEMIN5 | Gem nuclear organelle associated protein 5 |
| IFIT5 | Interferon induced protein with tetratricopeptide repeats 5 |
| LARP1 | La ribonucleoprotein 1, translational regulator |
| LSM1 | LSM1 homolog, mRNA degradation-associated |
| METTL1 | Methyltransferase 1, tRNA methylguanosine |
| NCBP1 | Nuclear cap binding protein subunit 1 |
| NCBP2 | Nuclear cap binding protein subunit 2 |
| NCBP2L | Nuclear cap binding protein subunit 2 like |
| NCBP3 | Nuclear cap binding protein subunit 3 |
| NSUN2 | NOP2/Sun RNA methyltransferase 2 |
| NUDT10 | Nudix hydrolase 10 |
| NUDT11 | Nudix hydrolase 11 |
| NUDT16 | Nudix hydrolase 16 |
| NUDT3 | Nudix hydrolase 3 |
| NUDT4 | Nudix hydrolase 4 |
| NUDT4B | Nudix hydrolase 4B |
| SNUPN | Snurportin 1 |
| WDR4 | WD repeat domain 4 |
m7G, N7-methylguanosine.
Data analysis
First, the differential expression of all m7G methylation-related genes between LSCC and adjacent normal tissues was determined, and then the data were imported into the limma R package (R Foundation for Statistical Computing, Vienna, Austria), according to a previous study (31). The genes with a log fold change (FC) >0.5 and an adjusted P value <0.001 were defined as DEGs.
Survival analysis and construction of a risk prediction model
First, we associated these 29 m7G methylation-related genes with the survival of LSCC patients using univariate Cox regression survival analysis. Second, we selected genes with a significant P value. The selections were instrumental to construct the risk prediction model by integrating the expression level of each gene and their corresponding coefficients. Third, we defined the risk prediction score as the risk score, and the predictive power of the risk score was used to predict the 1-, 2-, and 3-year survival rates of the patients using receiver operating characteristic (ROC) curve analysis. Finally, we further performed univariate and multivariate Cox regression analyses to identify the most significant independent risk factors for LSCC patients.
Construction of a protein-protein interaction (PPI) network
As the functions of any given gene are mediated through their coding proteins, a PPI network of the 29 m7G methylation-related genes using the igraph package in R was constructed. The PPI network shows the 29 m7G methylation-related genes as nodes, whereas each line connecting two nodes illustrates their biological relationship. Red lines indicate an upregulated correlation, whereas blue lines a downregulated correlation, with the color intensity representing the strength of the correlation.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses
Next, we performed GO and KEGG pathway enrichment analyses of the DEGs to identify their interactions and gene pathways in LSCC development. In brief, based on the risk score (see above), we categorized all patients into a high- or low-risk category using the median risk score as the cut-off value and utilized the limma R package to identify all DEGs between the high- and low-risk categories. Genes were defined as DEGs when the false discovery rate (FDR) <0.05 and |log2FC| ≥1, and then these genes were analyzed for the GO terms and KEGG pathways using the clusterProfiler package in R, according to a previous study (32).
Numeration of the immune infiltration score
The immune response is important in NSCLC, especially in tobacco smoke-related carcinogenesis (33,34). Thus, we profiled the immune cell population and activation of immune-associated pathways in the tumor microenvironment, gene set signatures in different immune cells, and immune-associated pathways from the literature (28) and utilized the molecular signature database to explore their role in LSCC (https://www.gsea-msigdb.org/gsea/msigdb/). The enrichment score of each signature for each LSCC sample was then inferred based on the RNA-sequencing data and the single-sample GSEA (ssGSEA) by using the gene set variation analysis (GSVA) R package.
Statistical analysis
In this study, inlimma R Software package and GraphPad Prism 5.0 (GraphPad Software Inc., La Jolla, CA, USA) were used for statistical analysis. T-test was used for comparison among groups difference analysis, ANOVA was used for continuous variables, and categorical variables were analyzed using Chi-square tests. P value less than 0.05 is defined as significance.
Results
Identification of differentially expressed m7G methylation-related genes in LSCC
In this study, the flow chart is shown in Figure 1, we first searched the TCGA database for DEGs in 502 LSCC samples versus 49 normal tissue samples and found 17 m7G methylation-related DEGs in LSCC tissues versus normal tissues using the following cut-off values: |logFC| >0.5, FDR <0.05, and P<0.01 (Figure 2A). Of these, 14 genes (NUDT10, NUDT3, NUDT11, SNUPN, AGO2, WDR4, DCPS, NCBP1, METTL1, LSM1, NSUN2, LARP1, NCBP2, and EIF3D) were upregulated in LSCC tissues, whereas 3 genes (NCBP2L, IFIT5, and EIF4E3) were downregulated. We then performed the PPI network analysis and evaluated the relationship among 29 m7G methylation-related genes (Figure 2B).


Classification and prognosis prediction of LSCC patients according to the expression of m7G methylation-related genes
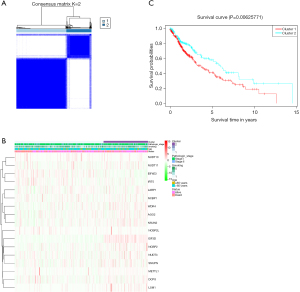
We analyzed the consensus clustering of the investigated 29 m7G-related genes in LSCC tissue samples by selecting k=2 to divide these 502 LSCC patients into high- and low-expression clusters (Figure 3A). The relationships between the gene expression profile and the clinical features (age ≤60 vs. >60 years), TNM stage, history of tobacco smoking, and survival data according to the high- and low-expression clusters) were presented in a heatmap (Figure 3B). Although we were unable to find significant differences in the clinical features between these two clusters, we did find a significant difference in the overall survival (OS) of patients between the two clusters (P=0.0062; Figure 3C), indicating the usefulness of this gene signature in predicting the prognosis of LSCC patients.
Univariate Cox regression analysis was used for primary screening of the survival-related genes. The results revealed that the expression of five m7G methylation-related genes (EIF3D, LSM1, NCBP2, NUDT10 and NUDT11) was significantly associated with the OS of the LSCC patients (Figure 4A). We then performed the least absolute shrinkage and selection operator (LASSO) Cox regression analysis to construct a gene signature using these five genes with the optimum λ value (Figure 4B). Subsequently, multivariate Cox regression analysis was performed to calculate the risk score for each LSCC case. The risk model formula for the risk score was as follows: risk score = (−0.005762 × EIF3D exp.) + (−0.007321 × LSM1 exp.) + (−0.005766 × NCBP2 exp.) + (−0.071864 × NUDT10 exp.) + (−0.012726 × NUDT11 exp.). We then classified these patients into high- and low-risk groups based on the cut-off value of the median risk score (Figure 4C) and performed principal component analysis (PCA; Figure 4D). We found that the LSCC patients with a high-risk score had a significantly shorter OS than those with a low-risk score (P=0.002; Figure 4E). Moreover, our ROC analysis revealed that such a risk score was able to predict the 1-, 2-, and 3-year survival rates of LSCC patients after surgery, with area under the curve (AUC) values of 0.542, 0.623, and 0.626, respectively (Figure 4F).

Identification of the signature of five m7G methylation-related genes as an independent prognostic factor for LSCC patients
Using univariate Cox regression analysis we stated that the higher the risk score was, the worse the prognosis of LSCC [hazard ratio (HR) =1.5503; 95% confidence interval (CI): 1.1585–2.0745; P=0.0032, Figure 5A]. To confirm the predictive value of this signature of five m7G methylation-related genes as an independent prognostic factor for LSCC patients, we performed a multivariate Cox regression analyses of the risk score and clinical features of the patients. We found that the risk score was an independent prognostic factor for the LSCC patients, specifically, the higher the risk score was, the worse the prognosis of LSCC (HR =1.57; 95% CI: 1.1702–2.1064; P=0.0026, Figure 5B). The pathological stage and the smoking status also emerged as independent factors for OS.

Furthermore, we associated this risk score with the clinicopathological features of the patients and the expression of the individual genes and found that the patients with a high-risk score exhibited significantly lower expression of NUDT10, NUDT11, LSM1, EIF3D, and NCBP2 (Figure 5C).
Association of the risk score with other biological features of LSCC
According to the risk score, we classified the patients and imported the data into the limma R package to search for the DEGs between the high- and low-risk groups using FDR <0.05 and |log2FC| ≥1. We obtained a total of 496 DEGs (332 upregulated genes and 164 downregulated genes in the high-risk group of patients; Table S1). We then performed GO and KEGG pathway analyses using the clusterProfiler package in R. Our data showed that the DEGs based on the risk model of this signature of five m7G methylation-related genes were mainly correlated with humoral immune response, cellular calcium ion homeostasis, and cytokine-cytokine receptor interaction (Figure 6A,6B).

Comparison of the immune activity between subgroups
To evaluate the changes in the immune profile according to the risk score of each patient, we enumerated the abundance of different tumor-infiltrating immune cells in the tumor mass using the whole-exome expression data (28) and found that the LSCC patients with high-risk scores had a low immune inflammatory microenvironment, which was shown by significantly lower levels of tumor-infiltrating immune cells such as T- and B-lymphocytes, dendritic cells, macrophages, and neutrophils (Figure 7A). After that, we obtained the enrichment scores for 13 immune-associated pathways, such as the cytotoxic activity, antigen presentation, inflammation- promoting, and interferon pathways, to compare their activities between the high- and low-risk cohorts of patients. As shown in Figure 7B, the high-risk-scored tumors had significantly lower activations of the pathways related to immune checkpoint activation, cytotoxic activity, antigen presentation, inflammatory response, or type II interferon response.

Discussion
LSCC treatment depends on many factors including the tumor stage, resectability, performance status, and genomic alterations; advanced unresectable LSCC tissues are treated with chemotherapy, radiation therapy, epidermal growth factor receptor (EGFR)-targeted therapy, antiangiogenetic therapy, and/or immune therapy; however, although the decades-long development of vascular endothelial growth factor receptor (VEGFR) inhibition and recent immunotherapy have been shown to improve the survival and quality of life of LSCC patients, the effectiveness of LSCC treatment has plateaued for several decades (35,36). At present, predictive biomarkers have been reported in LUAD (37,38), few studies was done in LSCC biomarkers. Thus, it is necessary to search for novel biomarkers could help medical oncologists to successfully predict LSCC treatment outcomes and prognoses. In the current study, we evaluated the prognostic significance of m7G methylation-related gene expression in LSCC tissues and established a signature of five m7G methylation-related genes (EIF3D, LSM1, NCBP2, NUDT10, and NUDT11) as a risk model to predict the OS of LSCC patients. We found that a high-risk score indicated a worse prognosis for LSCC patients. We also demonstrated that LSCC tissues from patients with a high-risk score were enriched with altered gene pathways that were related to the immune response and inflammatory response.
To the best of our knowledge, this is one of only a few studies of m7G methylation in lung cancer available in the literature (23) that explores the usefulness of m7G methylation-related genes as a predictive marker in LSCC. In the current study, we analyzed 29 m7G methylation-related genes in LSCC tissues versus normal specimens and identified five m7G methylation-related genes as a risk model to estimate the survival of LSCC patients. Specifically, eukaryotic translation initiation factor 3 (EIF3) is necessary for the initiation of protein synthesis in cells and consists of subunits of EIF3A-M; if its subunits are altered, oncogene expression is upregulated, and tumor transformation occurs (39-41). For example, EIF3D, as the core subunit of EIF3, plays an oncogenic role in NSCLC (42), and the percentage of apoptotic cells in renal cell carcinoma cells was increased after knockdown of EIF3D expression (43). The U6 small nuclear RNA (snRNA)-associated Sm-like protein LSM1 functions to intimately connect with RNA processing and degradation (44-47). In addition, the downregulation of LSM1 expression is associated with breast cancer progression (48,49). Moreover, nuclear cap-binding proteins 2 (NCBP2) encodes a subunit of the nuclear cap-binding complex to directly contact the 5'-cap through the RNA recognition motif (50) by binding to the 7-methylauanosin cap and is then added to the emerging 5'-end of nascent RNA to protect against 5'–3' exonucleolysis (51). A previous study has shown that NCBP2 can mediate gene interactions to modify neurodevelopmental defects of the 3q29 deletion (52). Additionally, nudix hydroxylases (NUDTs) belong to the versatile, widely distributed, housecleaning enzyme family to catalyze the hydrolysis of a wide range of nucleoside pyrophosphates linked to amino acids (53,54). Furthermore, previous researches have reported that NUDT10 expression is associated with malignant behaviors of gastric cancer by promoting tumor cell invasion and a poor prognosis (55,56), whereas NUDT11 has been shown to be associated with longer survival period in liver cancer (57) and bladder cancer (58). In our current study, we revealed that the expression of these five m7G methylation-related genes was associated with a poor prognosis of LSCC patients. However, further investigation is needed to understand their role in LSCC development and progression.
As PPI measurements have increased, more and more PPI network-based protein function prediction methods have been proposed and are generally superior to the homology-based prediction methods (59). We analyzed the PPI among 29 m7G methylation-related genes, and found that LARP1, DCP2 and GEMIN5 have the strongest positive relevance (the lines among three genes are the reddest). Although the genes regulating the m7G process have been found involved in the carcinogenesis process, the potential modulation between tumor immunity and m7G methylation-related genes remains elusive. Based on the DEGs between different risk groups, we performed GO analyses and discovered that humoral immune responses were enriched. To further explore the correlation between the risk score and immune status, we profiling immune cell populations, immune-associated pathways, and the role of gene set signatures with ssGSEA. Interestingly, we found that the high-risk groups have higher fractions of immune activity. The reason maybe was excessive immune activation promotes immune invasion, and this phenomenon has been found in hepatocellular carcinoma (60,61).
Despite our current data being interesting and potentially useful for determining the prognosis of LSCC patients, this study does have its limitations. For example, we only analyzed the data from the public TCGA database for construction of the risk model for LSCC. The expression of these genes at the protein level needs to be determined to clarify their association with LSCC prognosis. In addition, our current study had no opportunity to validate the expression, functions, and mechanisms of these genes using our own cohort of tissue specimens and cell lines.
Conclusions
The data from the current study demonstrated that detection of the signature of five m7G methylation-related genes could be useful for predicting the prognosis of LSCC patients. Moreover, this five-gene signature is an independent prognostic predictor for the survival of LSCC patients. Our future study will provide validation of our current data using a collection of LSCC samples.
Acknowledgments
Funding: This study was supported in part by a grant from
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1504/rc
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1504/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1504/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Dietel M, Bubendorf L, Dingemans AM, et al. Diagnostic procedures for non-small-cell lung cancer (NSCLC): recommendations of the European Expert Group. Thorax 2016;71:177-84. [Crossref] [PubMed]
- Balata H, Fong KM, Hendriks LE, et al. Prevention and Early Detection for NSCLC: Advances in Thoracic Oncology 2018. J Thorac Oncol 2019;14:1513-27. [Crossref] [PubMed]
- Omare MO, Kibet JK, Cherutoi JK, et al. A review of tobacco abuse and its epidemiological consequences. Z Gesundh Wiss 2022;30:1485-500. [Crossref] [PubMed]
- McRobbie H, Kwan B. Tobacco use disorder and the lungs. Addiction 2021;116:2559-71. [Crossref] [PubMed]
- Liu B, Liu Y, Zou J, et al. Smoking is Associated with Lung Adenocarcinoma and Lung Squamous Cell Carcinoma Progression through Inducing Distinguishing lncRNA Alterations in Different Genders. Anticancer Agents Med Chem 2022;22:1541-50. [Crossref] [PubMed]
- Kenfield SA, Wei EK, Stampfer MJ, et al. Comparison of aspects of smoking among the four histological types of lung cancer. Tob Control 2008;17:198-204. [Crossref] [PubMed]
- Gómez-López S, Whiteman ZE, Janes SM. Mapping lung squamous cell carcinoma pathogenesis through in vitro and in vivo models. Commun Biol 2021;4:937. [Crossref] [PubMed]
- Pan Y, Han H, Labbe KE, et al. Recent advances in preclinical models for lung squamous cell carcinoma. Oncogene 2021;40:2817-29. [Crossref] [PubMed]
- Dumitrescu RG. Epigenetic markers of early tumor development. Methods Mol Biol 2012;863:3-14. [Crossref] [PubMed]
- Brzeziańska E, Dutkowska A, Antczak A. The significance of epigenetic alterations in lung carcinogenesis. Mol Biol Rep 2013;40:309-25. [Crossref] [PubMed]
- Thapar R, Bacolla A, Oyeniran C, et al. RNA Modifications: Reversal Mechanisms and Cancer. Biochemistry 2019;58:312-29. [Crossref] [PubMed]
- Zhang LS, Liu C, Ma H, et al. Transcriptome-wide Mapping of Internal N(7)-Methylguanosine Methylome in Mammalian mRNA. Mol Cell 2019;74:1304-1316.e8. [Crossref] [PubMed]
- Tomikawa C. 7-Methylguanosine Modifications in Transfer RNA (tRNA). Int J Mol Sci 2018;19:4080. [Crossref] [PubMed]
- Kim SH, Suddath FL, Quigley GJ, et al. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science 1974;185:435-40. [Crossref] [PubMed]
- Jiao L, Liu Y, Yu XY, et al. Ribosome biogenesis in disease: new players and therapeutic targets. Signal Transduct Target Ther 2023;8:15. [Crossref] [PubMed]
- Jiapaer Z, Su D, Hua L, et al. Regulation and roles of RNA modifications in aging-related diseases. Aging Cell 2022;21:e13657. [Crossref] [PubMed]
- Boccaletto P, Stefaniak F, Ray A, et al. MODOMICS: a database of RNA modification pathways. 2021 update. Nucleic Acids Res 2022;50:D231-5. [Crossref] [PubMed]
- Cui L, Ma R, Cai J, et al. RNA modifications: importance in immune cell biology and related diseases. Signal Transduct Target Ther 2022;7:334. [Crossref] [PubMed]
- Li J, Zhang H, Wang H N. (1)-methyladenosine modification in cancer biology: Current status and future perspectives. Comput Struct Biotechnol J 2022;20:6578-85. [Crossref] [PubMed]
- Mattay J. Noncanonical metabolite RNA caps: Classification, quantification, (de)capping, and function. Wiley Interdiscip Rev RNA 2022;13:e1730. [Crossref] [PubMed]
- Alexandrov A, Martzen MR, Phizicky EM. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA 2002;8:1253-66. [Crossref] [PubMed]
- Teng PC, Liang Y, Yarmishyn AA, et al. RNA Modifications and Epigenetics in Modulation of Lung Cancer and Pulmonary Diseases. Int J Mol Sci 2021;22:10592. [Crossref] [PubMed]
- Shaheen R, Abdel-Salam GM, Guy MP, et al. Mutation in WDR4 impairs tRNA m(7)G46 methylation and causes a distinct form of microcephalic primordial dwarfism. Genome Biol 2015;16:210. [Crossref] [PubMed]
- Cui W, Zhao D, Jiang J, et al. tRNA Modifications and Modifying Enzymes in Disease, the Potential Therapeutic Targets. Int J Biol Sci 2023;19:1146-62. [Crossref] [PubMed]
- Pandolfini L, Barbieri I, Bannister AJ, et al. METTL1 Promotes let-7 MicroRNA Processing via m7G Methylation. Mol Cell 2019;74:1278-1290.e9. [Crossref] [PubMed]
- Chen Y, Xie C, Zheng X, et al. LIN28/let-7/PD-L1 Pathway as a Target for Cancer Immunotherapy. Cancer Immunol Res 2019;7:487-97. [Crossref] [PubMed]
- Pan J, Huang Z, Lin H, et al. M7G-Related lncRNAs predict prognosis and regulate the immune microenvironment in lung squamous cell carcinoma. BMC Cancer 2022;22:1132. [Crossref] [PubMed]
- Qiu X, Chen Y, Zhu X, et al. Analysis and Validation of the Prognosis ability of the M7GRelated miRNAs in Lung Adenocarcinoma. Asian Pac J Cancer Prev 2023;24:1275-87. [Crossref] [PubMed]
- Tao X, Huang R, Xu R, et al. A novel m7G methylation-related signature associated with chromosome homeostasis in patients with lung adenocarcinoma. Front Genet 2022;13:998258. [Crossref] [PubMed]
- Liu S, Wang Z, Zhu R, et al. Three Differential Expression Analysis Methods for RNA Sequencing: limma, EdgeR, DESeq2. J Vis Exp 2021;
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Yuan H, Liu J, Zhang J. The Current Landscape of Immune Checkpoint Blockade in Metastatic Lung Squamous Cell Carcinoma. Molecules 2021;26:1392. [Crossref] [PubMed]
- Hendry S, Salgado R, Gevaert T, et al. Assessing Tumor-Infiltrating Lymphocytes in Solid Tumors: A Practical Review for Pathologists and Proposal for a Standardized Method from the International Immuno-Oncology Biomarkers Working Group: Part 2: TILs in Melanoma, Gastrointestinal Tract Carcinomas, Non-Small Cell Lung Carcinoma and Mesothelioma, Endometrial and Ovarian Carcinomas, Squamous Cell Carcinoma of the Head and Neck, Genitourinary Carcinomas, and Primary Brain Tumors. Adv Anat Pathol 2017;24:311-35. [Crossref] [PubMed]
- Sasa GBK, Xuan C, Chen M, et al. Clinicopathological implications of lncRNAs, immunotherapy and DNA methylation in lung squamous cell carcinoma: a narrative review. Transl Cancer Res 2021;10:5406-29. [Crossref] [PubMed]
- Tian Y, Zhai X, Yan W, et al. Clinical outcomes of immune checkpoint blockades and the underlying immune escape mechanisms in squamous and adenocarcinoma NSCLC. Cancer Med 2021;10:3-14. [Crossref] [PubMed]
- Tran TO, Vo TH, Lam LHT, et al. ALDH2 as a potential stem cell-related biomarker in lung adenocarcinoma: Comprehensive multi-omics analysis. Comput Struct Biotechnol J 2023;21:1921-9. [Crossref] [PubMed]
- Dang HH, Ta HDK, Nguyen TTT, et al. Prospective role and immunotherapeutic targets of sideroflexin protein family in lung adenocarcinoma: evidence from bioinformatics validation. Funct Integr Genomics 2022;22:1057-72. [Crossref] [PubMed]
- Yi SH, Petrychenko V, Schliep JE, et al. Conformational rearrangements upon start codon recognition in human 48S translation initiation complex. Nucleic Acids Res 2022;50:5282-98. [Crossref] [PubMed]
- Ma S, Liu JY, Zhang JT. eIF3d: A driver of noncanonical cap-dependent translation of specific mRNAs and a trigger of biological/pathological processes. J Biol Chem 2023;299:104658. [Crossref] [PubMed]
- Rubio A, Garland GD, Sfakianos A, et al. Aberrant protein synthesis and cancer development: The role of canonical eukaryotic initiation, elongation and termination factors in tumorigenesis. Semin Cancer Biol 2022;86:151-65. [Crossref] [PubMed]
- Wang D, Jia Y, Zheng W, et al. Overexpression of eIF3D in Lung Adenocarcinoma Is a New Independent Prognostic Marker of Poor Survival. Dis Markers 2019;2019:6019637. [Crossref] [PubMed]
- Chen M, Nie Z, Gao Y, et al. m7G regulator-mediated molecular subtypes and tumor microenvironment in kidney renal clear cell carcinoma. Front Pharmacol 2022;13:900006. [Crossref] [PubMed]
- Kok Kilic G, Isik E, Alpay O, et al. LSM1 is the new candidate gene for neurodevelopmental disorder. Eur J Med Genet 2022;65:104610. [Crossref] [PubMed]
- Zhu J, Chen K, Sun YH, et al. LSM1-mediated Major Satellite RNA decay is required for nonequilibrium histone H3.3 incorporation into parental pronuclei. Nat Commun 2023;14:957. [Crossref] [PubMed]
- He F, Jacobson A. Eukaryotic mRNA decapping factors: molecular mechanisms and activity. FEBS J 2023;290:5057-85. [Crossref] [PubMed]
- Voutsadakis IA. Characteristics and Prognosis of 8p11.23-Amplified Squamous Lung Carcinomas. J Clin Med 2023;12:1711. [Crossref] [PubMed]
- Tzeng YT, Tsui KH, Tseng LM, et al. Integrated analysis of pivotal biomarker of LSM1, immune cell infiltration and therapeutic drugs in breast cancer. J Cell Mol Med 2022;26:4007-20. [Crossref] [PubMed]
- Voutsadakis IA. 8p11.23 Amplification in Breast Cancer: Molecular Characteristics, Prognosis and Targeted Therapy. J Clin Med 2020;9:3079. [Crossref] [PubMed]
- Calero G, Wilson KF, Ly T, et al. Structural basis of m7GpppG binding to the nuclear cap-binding protein complex. Nat Struct Biol 2002;9:912-7. [Crossref] [PubMed]
- Pabis M, Neufeld N, Shav-Tal Y, et al. Binding properties and dynamic localization of an alternative isoform of the cap-binding complex subunit CBP20. Nucleus 2010;1:412-21. [Crossref] [PubMed]
- Singh MD, Jensen M, Lasser M, et al. NCBP2 modulates neurodevelopmental defects of the 3q29 deletion in Drosophila and Xenopus laevis models. PLoS Genet 2020;16:e1008590. [Crossref] [PubMed]
- Gong J, Yang J, He Y, et al. Construction of m7G subtype classification on heterogeneity of sepsis. Front Genet 2022;13:1021770. [Crossref] [PubMed]
- Malik A, Hept MA, Purcell EB. Sound the (Smaller) Alarm: The Triphosphate Magic Spot Nucleotide pGpp. Infect Immun 2023;91:e0043222. [Crossref] [PubMed]
- Li XY, Wang SL, Chen DH, et al. Construction and Validation of a m7G-Related Gene-Based Prognostic Model for Gastric Cancer. Front Oncol 2022;12:861412. [Crossref] [PubMed]
- Chen D, Zhang R, Xie A, et al. Clinical correlations and prognostic value of Nudix hydroxylase 10 in patients with gastric cancer. Bioengineered 2021;12:9779-89. [Crossref] [PubMed]
- Ren S, Cao W, Ma J, et al. Correlation evaluation between cancer microenvironment related genes and prognosis based on intelligent medical internet of things. Front Genet 2023;14:1132242. [Crossref] [PubMed]
- Li DX, Feng DC, Wang XM, et al. M7G-related molecular subtypes can predict the prognosis and correlate with immunotherapy and chemotherapy responses in bladder cancer patients. Eur J Med Res 2023;28:55. [Crossref] [PubMed]
- Wu Z, Liao Q, Liu B. A comprehensive review and evaluation of computational methods for identifying protein complexes from protein-protein interaction networks. Brief Bioinform 2020;21:1531-48. [Crossref] [PubMed]
- Zhang Q, He Y, Luo N, et al. Landscape and Dynamics of Single Immune Cells in Hepatocellular Carcinoma. Cell 2019;179:829-845.e20. [Crossref] [PubMed]
- Zhou SL, Zhou ZJ, Hu ZQ, et al. Tumor-Associated Neutrophils Recruit Macrophages and T-Regulatory Cells to Promote Progression of Hepatocellular Carcinoma and Resistance to Sorafenib. Gastroenterology 2016;150:1646-1658.e17. [Crossref] [PubMed]


