
Genomic profiling of relapsed small cell lung cancer reveals potential pathways of therapeutic targets
Highlight box
Key findings
• It was identified several genomic alterations with potential therapeutic implications for small cell lung cancer (SCLC).
• DNA damage response pathways represented attractive targets in relapsed SCLC.
• Genotyping of relapsed SCLC may reveal patient who has benefit from targeted therapy.
What is known and what is new?
• SCLC is characterized by high growth fraction, rapid doubling time, limited treatment options, and poor clinical outcomes. Previous studies have revealed a few recurrently mutated genes in untreated SCLC, whereas the genomic profiling of relapsed SCLC remains unclear.
• Our study first exhibited comprehensive genomic profiling of relapsed SCLC, identifying several candidate genes for exploring potential targets or biomarkers.
What is the implication, and what should change now?
• Next-generation sequencing for monitoring relapsed SCLC evolution is essential to develop more effective therapeutic options and potential biomarkers.
Introduction
Small cell lung cancer (SCLC), which accounts for approximately 15% of lung cancer cases, is characterized by high growth fraction, rapid doubling time, and poor clinical outcomes (1). Despite the past 30 years of clinical trials aimed at improving the outcomes for SCLC, the majority of SCLC remains incurable (2). Currently, chemotherapy remains the cornerstone of therapy for both extensive disease (ED) and limited disease (LD)-SCLC, with high response rates to first-line cytotoxic chemotherapy; the response rates of first-line platinum plus etoposide (EP) chemotherapy are 50–70% (2,3). However, in most cases, the duration of the clinical response is extremely limited, with nearly all patients relapsing, usually within a few months (1,4-6). Unfortunately, therapeutic options available for relapsed SCLC patients are limited: the response rates to second-line chemotherapy are as low as 5–30% (7-11); and there are currently no United States Food and Drug Administration (FDA)-approved targeted agents for the treatment of SCLC; immune checkpoint inhibitors (ICIs), as the first non-chemotherapy alternative for patients with metastatic and relapsed SCLC (2,11-14), are of benefit to only a minority of patients, who cannot be well identified by existing biomarkers. Therefore, there is an urgent need to explore more effective therapies and specific biomarkers in relapsed SCLC.
Genetic testing could be used to better understand the pathogenesis of SCLC and to explore the potential pathways or targets for therapeutic strategies. Previous study has revealed a few recurrently mutated genes in untreated SCLC (15), whereas the genomic profiling of relapsed SCLC remains unclear, and there is a lack of data on the impact of genomic alterations on therapeutic options and survival among patients with relapsed SCLC. To develop more effective therapeutic strategies and optimal biomarkers of SCLC, it is necessary to improve our understanding of genomic profiling of relapsed SCLC.
Consequently, a consecutive cohort of 31 patients, diagnosed with relapsed SCLC in Zhongnan Hospital, Wuhan University between October 2018 and November 2019, was recruited and followed up. Blood samples were collected at enrollment for next-generation sequencing (NGS), to analyze the comprehensive genomic profiling of relapsed SCLC, and to explore the impact of mutated genes on therapeutic options and survival among patients with relapsed SCLC. We aimed to yield instructive results for the development of innovative clinical management strategies and specific biomarkers for patients with SCLC. We present this article in accordance with the REMARK reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1657/rc).
Methods
Patients
Between October 2018 and November 2019, there were a total of 68 consecutive patients with SCLC at the Department of Pulmonary Oncology, Zhongnan Hospital, Wuhan University. Among the 68 patients, a total of 31 eligible patients with relapsed SCLC were enrolled in this study. Thirty-seven patients of those were excluded due to non-relapsed SCLC. Our inclusion criteria were as follows: (I) SCLC confirmed by cytology (2 patients), or histology (29 patients) [World Health Organization (WHO)]; (II) the patients diagnosed with relapsed SCLC, who had undergone at least the first-line chemotherapy. The whole blood specimens from 31 patients were collected for NGS assessment (Euler Genomics Technology Co., Ltd., Beijing, China) at enrollment, circulating tumor DNA (ctDNA) extracted from the plasma separated from whole blood specimen was sequenced to high, uniform coverage (Illumina HiSeq 2500; Illumina, San Diego, CA, USA) and analyzed for all classes of genomic alterations. Genomic alterations of these patient samples, including base substitutions, small indels, rearrangements, copy number alterations, and so on, were determined and then reported. Additionally, the clinical characteristics and follow-up information of included patients were collected and analyzed.
The study was approved by the Ethical Committee of Zhongnan Hospital, Wuhan University (No. 2021017). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Written informed consent was provided by all included patients. Additionally, blood samples from participants with relapsed SCLC were collected for free NGS assessment, the results of which might help physicians to make decisions for subsequent treatment. We collected participant information by reviewing their electronic medical records, for which we preserved patient anonymity. We ensured that the data collection, analysis, and publication would not jeopardize participants’ health, safety, or privacy.
Treatment and follow-up
All 31 patients with relapsed SCLC had received EP as the first-line chemotherapy. Blood samples were collected for NGS assessment in 16 patients who exhibited disease progression following the first-line treatment, in 13 patients with disease progression following the second-line treatment, and in 2 patients with disease progression following the third-line treatment.
Follow-up examinations were performed every 2 months, including thoracic and abdominal computed tomography (CT) scan, and brain magnetic resonance imaging (MRI) scan. Progression-free survival (PFS) was defined as the duration from the onset of illness to the failure of the first-line treatment. Overall survival (OS) was defined as the duration from the onset of illness to death from any cause.
Statistics
All statistical analyses were performed by using SPSS 22.0 (IBM Corp., Armonk, NY, USA). Descriptive statistics are used for categorical variables (frequency and percentage) and continuous variables (median and range). The cumulative survival rate was calculated by the Kaplan-Meier method with 95% confidence intervals (CIs). Univariable Cox regression analyses was used to investigate the factors associated with PFS and OS. All tests were two-sided and P<0.05 was considered statistically significant.
Results
Patient characteristics and treatment
Between October 2018 and November 2019, a total of 31 eligible patients with relapsed SCLC were analyzed in the current study. The baseline characteristics of these patients are shown in Table 1. In detail, there were 27 male and 4 female relapsed SCLC patients with a median age of 62 (range, 43–75) years. The majority of patients (20 patients, 64.5%) were heavy smokers, and 96.8% of patients (30 patients) presented a Karnofsky performance status (KPS) score ≥80. There were 15 patients of LD stage and 16 patients of ED stage. Of the 15 LD stage patients, 3 cases received surgery initially. All patients had received prior chemotherapy and had disease progression at enrollment. The number of prior lines of cancer treatment ranged from 1 to 3 (median 2) before NGS assessment, and 64.5% (20 patients) were platinum sensitive, defined here as having a chemotherapy-free interval of at least 90 days from completion of the first-line treatment to initiation of the second-line treatment. Additionally, all of 31 patients received radiotherapy during the whole course of treatment, whereas before NGS sequencing, a total of 3 patients did not receive radiotherapy.
Table 1
| Characteristics | Value (n=31) |
|---|---|
| Age (years) | 62 [43–75] |
| ≥60 | 19 (61.3) |
| <60 | 12 (38.7) |
| Gender | |
| Male | 27 (87.1) |
| Female | 4 (12.9) |
| KPS score | |
| ≥80 | 30 (96.8) |
| <80 | 1 (3.2) |
| Smoking history | |
| Yes | 20 (64.5) |
| No | 11 (35.5) |
| Clinical stages | |
| LD | 15 (48.4) |
| ED | 16 (51.6) |
| Pattern of relapse after the first-line chemotherapy | |
| Platinum-sensitive disease (progression ≥90 days) | 20 (64.5) |
| Platinum-refractory disease (progression <90 days) | 11 (35.5) |
| Whether patients received radiotherapy before NGS sequencing? | |
| Yes | 28 (90.3) |
| No | 3 (9.7) |
| Prior lines of therapy before NGS sequencing | 2 [1–3] |
| 1 | 16 (51.6) |
| 2 | 13 (41.9) |
| 3 | 2 (6.5) |
| Time from diagnosis to DNA sequencing (months) | 10.9 [2.1–47.6] |
| TMB | 6 [1–66] |
| Low, TMB <20 | 26 (83.9) |
| Medium, 20≤ TMB <40 | 3 (9.7) |
| High, 40≤ TMB <60 | 1 (3.2) |
| Very high, TMB ≥60 | 1 (3.2) |
| MSI/MSS | 12 [4.1–32.0] |
| Stable (MSS), MSI <20 | 27 (87.1) |
| Moderate instable (MSI), 20≤ MSI <40 | 4 (12.9) |
| High instable (MSI-H), 40≤ MSI <60 | 0 (0.0) |
| Very high instable (MSI-VH), MSI ≥60 | 0 (0.0) |
| Genomic alterations per sample | 5 [1–11] |
Values are presented as median [range] or n (%). KPS, Karnofsky performance status; LD, limited disease; ED, extensive disease; NGS, next-generation sequencing; TMB, tumor mutation burden; MSI, microsatellite instability; MSS, microsatellite stability; MSI-H, MSI-high; MSI-VH, MSI-very high.
The comprehensive genomic profiling of relapsed SCLC
Among the 31 blood samples used for DNA extraction, no differences were observed in the quality of sequencing results. All 31 relapsed SCLC specimens (100%) harbored at least 1 genomic alteration, with 162 total alterations identified for a median of 5 alterations per sample (range, 1–11). Additionally, patients with relapsed SCLC exhibited relatively low blood tumor mutation burden (TMB), among whom most patients (26 patients, 83.9%) were defined as low TMB (TMB value <20) according to the cut-off value of TMB in this current NGS sequencing package. Likewise, the majority of patients (27 patients, 87.1%) were defined as microsatellite stability (MSS) [microsatellite instability (MSI) value <20] (Table 1).
Consistent with prior reports of untreated SCLC (15,16), the most frequent genomic alterations observed in relapsed SCLC were also TP53 (24 patients, 77%) and RB1 (16 patients, 52%) mutations. Other genomic alterations of high frequency were breast cancer 2 (BRCA2) (10 patients, 32%), ataxia telangiectasia mutated (ATM) (4 patients, 13%), epidermal growth factor receptor (EGFR) (3 patients, 10%), Notch receptor 1 (NOTCH1) (3 patients, 10%), Fanconi anemia complementation group A (FANCA) (3 patients, 10%) in turn (Figure 1). It was worth highlighting that untreated SCLC was not characterized by frequent mutations in BRCA1/2 (15-17), which was not consistent with relapsed SCLC in our current study.

The survival of relapsed SCLC cohort
For the cohort of relapsed SCLC patients, the median duration of follow-up was 36.5 months (95% CI: 10.4–62.6). The median OS of these patients was 17.8 months (95% CI: 16.1–19.5) (Figure 2). The 1- and 2-year OS rates were 67.7% and 32.0%, respectively. For PFS, all patients had disease progression and had received a least the first line of chemotherapy at enrollment. Thus, the PFS of first-line treatment was defined as the duration from the onset of illness to the failure of the first-line treatment. The median PFS was 7.9 months (95% CI: 6.2–9.7) (Figure 2). The 1- and 2-year PFS rates were 25.8% and 12.9% respectively. At last, the median duration from the onset of illness to enrollment/NGS assessment was 10.9 months. As expected, the clinical stages and pattern of relapse were confirmed significantly associated with OS and PFS by univariable Cox regression analyses (P<0.05).

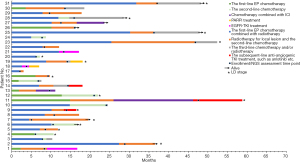
The individualized approach and survival of relapsed SCLC cohort
Moreover, we analyzed the impact of genomic alterations on therapeutic options and survival among patients with relapsed SCLC. The individualized approach and survival of 31 relapsed SCLC patients are shown in Figure 3. A total of 3 patients (No. 11, 13, and 27) eventually received ICIs-based treatment as the third-line therapy, and their PFS of ICIs-based treatment was 20.3, 4.9, and 7.0 months, respectively. Interestingly, all of these 3 cases exhibited low TMB and MSS. For patients with mutations of potential therapeutic targets, cases No. 18 and 19 had mutation in BRCA2, were treated with poly-ADP-ribose polymerase inhibitor (PARPi, olaparib) monotherapy in the subsequent-line therapy, whereas the PFS was 2.53 and 3.1 months, respectively. In addition, there was 1 male (No. 18) and 2 females (No. 1 and 21) harbored common EGFR mutation. In detail, 19-del mutation in case No. 18, and L858R mutation in cases No. 1 and 21. After EGFR tyrosine kinase inhibitor (TKI) treatment, the male patient exhibited a PFS of only 1.87 months, and the PFS of the 2 females were 5.1 and 7.7 months, respectively. At last, we observed that a total of 3 patients with absence of driver mutations (No. 9, 11, and 14) receiving anti-angiogenic TKI treatment showed the PFS of 3.2, 12.53, and 3.7 months, respectively.
Discussion
SCLC remains one of the most aggressive and lethal malignancies, with a minimal role of curative surgery and no targetable agents approved. Almost all patients with SCLC relapse ultimately. Over the past two decades, there has been no significant improvement in clinical outcomes for patients with SCLC. Although ICIs were demonstrated as the first and only non-chemotherapy option with survival benefit to SCLC, only a minority of SCLC patients could benefit from immunotherapy, and there was a lack of the specific biomarker for identification optimal sub-population that could benefit from immunotherapy (2,11-14). Consequently, exploring the genomic profiling of relapsed SCLC could help to develop more effective therapeutic options and to explore optimal biomarkers. A recent consensus proposal suggested grouping SCLC into four subtypes defined by expression of ASCL1, NEUROD1, POU2F3, and YAP1, referring to these respectively as SCLC-A, SCLC-N, SCLC-P, and SCLC-Y. Whereas the clinical implications of this molecular categorization remain to be explained (18). Recently, the liquid biopsy in genetic sequencing could potentially allow physicians to identify patients whose tumors harbor specific mutations in the least invasive way. Considering that repeated biopsies after initial treatment are typically outside of standard of care in SCLC, genomic profiling of relapsed SCLC in the current study was detected by blood sample.
The median OS and the first-line treatment PFS of our relapsed SCLC cohort was 17.8 and 7.9 months (Figure 2), respectively. As expected, the clinical stages and pattern of relapse (platinum-sensitive or -refractory) were confirmed as significantly associated with PFS and OS (P<0.05), whereas it was more concerned with the significance of genomic alterations on therapeutic options and survival among relapsed SCLC. As in previous reports, the absolute number of genomic alterations in most SCLC patients is much higher than in other tumors (15,16,19). In our cohort of relapsed SCLC, the median number of genomic alterations per sample was 5 (range, 1–11). Whereas in contrast to untreated SCLC previously reported (15,16,20), patients with relapsed SCLC exhibited relatively low TMB. The majority of patients (26 patients, 83.9%) were defined as low TMB (TMB value <20) according to the cut-off value of TMB in the current NGS sequencing package. Likewise, most patients (27 patients, 87.1%) were defined as MSS (MSI value <20). Recent studies have indicated that TMB or MSI might be a useful biomarker to predict likelihood of response to ICIs treatment among SCLC (21-23). Therefore, the above characteristics mentioned of relapsed SCLC might imply non-responsive to ICIs treatment. However, we observed that the ICIs-based treatment still brought considerable PFS (4.93–20.27 months) for 3 patients (No. 11, 13, and 27) with low TMB and MSS. Therefore, it is arguable that TMB or MSI is a useful biomarker for ICIs treatment among SCLC. Our findings also suggested the need for the development of optimal biomarker in further research.
Moreover, the most frequent genetic alterations observed in relapsed SCLC were TP53 (77%) and RB1 (52%) mutations, which was consistent with previous reports of untreated SCLC (15,16). Our results further provide evidence for universal bi-allelic inactivation of TP53 and RB1 in SCLC, thereby reconfirming the two genes as obligatory tumor suppressors of SCLC. TP53 and RB1 control and influence most of the important pathways involved in detecting DNA damage, regulating cell cycle, and inducing apoptosis (24). As all control of the main tumor suppressive pathways is lost, SCLC shows the quickest and most aggressive growth among all tumor entities. Additionally, SCLC is one of tumors with the best initial response to chemotherapy and radiotherapy, probably also because SCLC induces a loss of ability to detect and repair DNA damage. Our findings reconfirmed that DNA damage response (DDR) pathways represented attractive targets in SCLC.
Although genomic alterations in TP53 and RB1 cannot be suitable targets for treatment currently, we still found several genetic alterations in relapsed SCLC, such as mutations in EGFR, BRCA1/2, PTEN, PIK3CA, and so on (Figure 1), which were identified in other tumors with a potential therapeutic implication (25). To our knowledge, untreated SCLC is not characterized by homologous recombination (HR) deficiency or frequent mutations in BRCA1/2 (15-17), which is contrast to our results of relapsed SCLC in this current study. BRCA1 and 2 deficient cells are rendered defective in the ability to repair through HR and depend on error-prone non-homologous end joining (NHEJ) for DNA repair (23-29). PARP targeting is highly effective in the context of synthetic lethality in models with existing deficiency in DNA repair, such as deleterious mutations in BRCA1/2 (26,27). Thus, it could be speculated that PARP targeting might be an attractive therapy option for relapsed SCLC. The PFS of BRCA2 mutant patients (No. 18 and 19) treated with PARPi (olaparib) were 2.53 and 3.1 months, respectively. It seemed monotherapy of olaparib had limited efficacy in relapsed SCLC, even in BRCA2 mutant patients. Therefore, the addition of other agents to PARPi treatment is probably an area worthy of further investigation among SCLC. Literatures and our previous study have proved (30,31) that DDR inhibitor can synergize with ICIs via the cGas-STING-IFN stimulation, which makes them a promising option for recurrent SCLC with DDR vulnerabilities.
Other genomic alterations such as mutations in EGFR, PTEN, PIK3CA, and so on, were also found in other tumors with a potential therapeutic implication. Therefore, genomic profiling of patients with relapsed SCLC may reveal individual patients who might receive potential benefit from targeted therapeutic interventions. In this current study, 3 patients (No. 18, 1, and 21) with common EGFR mutation all received EGFR-TKI treatment. And none of these cases is transformation of EGFR-mutation adeno-carcinomas. The high rate of EGFR-mutation in our cohort is partly explained by a small sample study, which inevitably resulted in a selection bias. However, the PFS of EGFR-TKI treatment among these relapsed SCLC patients was significantly poorer than that of lung adenocarcinoma patients with EGFR mutation. It was indicated that several somatic mutations do not cause genomic dependence and do not cause vulnerability to targeted therapies in SCLC. Additionally, we observed 3 patients (No. 9, 11, and 14) receiving anti-angiogenic TKI treatment who showed the PFS of 3.2, 12.53, and 3.7 months, respectively, which indicated the role of anti-angiogenic TKI in subsequent-line treatment. Consequently, our findings indicated that high-sensitivity genomic profiling could reveal potential new pathways to targeted therapies among patients with SCLC who have relapsed after primary chemotherapy.
Conclusions
Our studies first exhibited comprehensive genomic profiling of relapsed SCLC, identifying several candidate genes, and briefly analyzed the association of survival and genomic alterations. Our data from a small cohort of relapsed SCLC will benefit further exploration the potential targets or biomarkers in relapsed SCLC. Certainly, there were several limitations in our study, it was a small sample study from a single institution, which inevitably resulted in a selection bias. More well-designed, prospective, and randomized studies are needed to confirm the conclusion. We anticipate that the next 20 years, more progress will be made in the treatment of SCLC than in the past, and that prognosis for patients with this disease will be greatly improved.
Acknowledgments
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1657/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1657/dss
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1657/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1657/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study and all experimental protocols were approved by the Ethics Committee of Zhongnan Hospital, Wuhan University (No. 2021017). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Written informed consent was provided by all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Liang J, Guan X, Bao G, et al. Molecular subtyping of small cell lung cancer. Semin Cancer Biol 2022;86:450-62. [Crossref] [PubMed]
- Horn L, Mansfield AS, Szczęsna A, et al. First-Line Atezolizumab plus Chemotherapy in Extensive-Stage Small-Cell Lung Cancer. N Engl J Med 2018;379:2220-9. [Crossref] [PubMed]
- Hiddinga BI, Raskin J, Janssens A, et al. Recent developments in the treatment of small cell lung cancer. Eur Respir Rev 2021;30:210079. [Crossref] [PubMed]
- Zugazagoitia J, Paz-Ares L. Extensive-Stage Small-Cell Lung Cancer: First-Line and Second-Line Treatment Options. J Clin Oncol 2022;40:671-80. [Crossref] [PubMed]
- Petty WJ, Paz-Ares L. Emerging Strategies for the Treatment of Small Cell Lung Cancer: A Review. JAMA Oncol 2023;9:419-29. [Crossref] [PubMed]
- Ganti AKP, Loo BW, Bassetti M, et al. Small Cell Lung Cancer, Version 2.2022, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2021;19:1441-64. [Crossref] [PubMed]
- Bernabé-Caro R, Chen Y, Dowlati A, et al. Current and Emerging Treatment Options for Patients With Relapsed Small-cell Lung Carcinoma: A Systematic Literature Review. Clin Lung Cancer 2023;24:185-208. [Crossref] [PubMed]
- Das M, Padda SK, Weiss J, et al. Advances in Treatment of Recurrent Small Cell Lung Cancer (SCLC): Insights for Optimizing Patient Outcomes from an Expert Roundtable Discussion. Adv Ther 2021;38:5431-51. [Crossref] [PubMed]
- Wang S, Zimmermann S, Parikh K, et al. Current Diagnosis and Management of Small-Cell Lung Cancer. Mayo Clin Proc 2019;94:1599-622. [Crossref] [PubMed]
- von Pawel J, Jotte R, Spigel DR, et al. Randomized phase III trial of amrubicin versus topotecan as second-line treatment for patients with small-cell lung cancer. J Clin Oncol 2014;32:4012-9. [Crossref] [PubMed]
- Antonia SJ, López-Martin JA, Bendell J, et al. Nivolumab alone and nivolumab plus ipilimumab in recurrent small-cell lung cancer (CheckMate 032): a multicentre, open-label, phase 1/2 trial. Lancet Oncol 2016;17:883-95. [Crossref] [PubMed]
- Paz-Ares L, Dvorkin M, Chen Y, et al. Durvalumab plus platinum-etoposide versus platinum-etoposide in first-line treatment of extensive-stage small-cell lung cancer (CASPIAN): a randomised, controlled, open-label, phase 3 trial. Lancet 2019;394:1929-39. [Crossref] [PubMed]
- Cheng Y, Han L, Wu L, et al. Effect of First-Line Serplulimab vs Placebo Added to Chemotherapy on Survival in Patients With Extensive-Stage Small Cell Lung Cancer: The ASTRUM-005 Randomized Clinical Trial. JAMA 2022;328:1223-32. [Crossref] [PubMed]
- Wang J, Zhou C, Yao W, et al. Adebrelimab or placebo plus carboplatin and etoposide as first-line treatment for extensive-stage small-cell lung cancer (CAPSTONE-1): a multicentre, randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2022;23:739-47. [Crossref] [PubMed]
- Peifer M, Fernández-Cuesta L, Sos ML, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 2012;44:1104-10. [Crossref] [PubMed]
- Sivakumar S, Moore JA, Montesion M, et al. Integrative Analysis of a Large Real-World Cohort of Small Cell Lung Cancer Identifies Distinct Genetic Subtypes and Insights into Histologic Transformation. Cancer Discov 2023;13:1572-91. [Crossref] [PubMed]
- Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet 2001;27:247-54. [Crossref] [PubMed]
- Baine MK, Hsieh MS, Lai WV, et al. SCLC Subtypes Defined by ASCL1, NEUROD1, POU2F3, and YAP1: A Comprehensive Immunohistochemical and Histopathologic Characterization. J Thorac Oncol 2020;15:1823-35. [Crossref] [PubMed]
- Iwakawa R, Kohno T, Totoki Y, et al. Expression and clinical significance of genes frequently mutated in small cell lung cancers defined by whole exome/RNA sequencing. Carcinogenesis 2015;36:616-21. [Crossref] [PubMed]
- Liu SV, Reck M, Mansfield AS, et al. Updated Overall Survival and PD-L1 Subgroup Analysis of Patients With Extensive-Stage Small-Cell Lung Cancer Treated With Atezolizumab, Carboplatin, and Etoposide (IMpower133). J Clin Oncol 2021;39:619-30. [Crossref] [PubMed]
- Ready NE, Ott PA, Hellmann MD, et al. Nivolumab Monotherapy and Nivolumab Plus Ipilimumab in Recurrent Small Cell Lung Cancer: Results From the CheckMate 032 Randomized Cohort. J Thorac Oncol 2020;15:426-35. [Crossref] [PubMed]
- Le DT, Kim TW, Van Cutsem E, et al. Phase II Open-Label Study of Pembrolizumab in Treatment-Refractory, Microsatellite Instability-High/Mismatch Repair-Deficient Metastatic Colorectal Cancer: KEYNOTE-164. J Clin Oncol 2020;38:11-9. [Crossref] [PubMed]
- Marabelle A, Fakih M, Lopez J, et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol 2020;21:1353-65. [Crossref] [PubMed]
- Emam M, Machado JP, Antunes A. Evolutionary genomics of mammalian lung cancer genes reveals signatures of positive selection in APC, RB1 and TP53. Genomics 2020;112:4722-31. [Crossref] [PubMed]
- Sharp A, Bhosle J, Abdelraouf F, et al. Development of molecularly targeted agents and immunotherapies in small cell lung cancer. Eur J Cancer 2016;60:26-39. [Crossref] [PubMed]
- Rose M, Burgess JT, O'Byrne K, et al. PARP Inhibitors: Clinical Relevance, Mechanisms of Action and Tumor Resistance. Front Cell Dev Biol 2020;8:564601. [Crossref] [PubMed]
- Dziadkowiec KN, Gąsiorowska E, Nowak-Markwitz E, et al. PARP inhibitors: review of mechanisms of action and BRCA1/2 mutation targeting. Prz Menopauzalny 2016;15:215-9. [Crossref] [PubMed]
- Lallo A, Frese KK, Morrow CJ, et al. The Combination of the PARP Inhibitor Olaparib and the WEE1 Inhibitor AZD1775 as a New Therapeutic Option for Small Cell Lung Cancer. Clin Cancer Res 2018;24:5153-64. [Crossref] [PubMed]
- Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005;434:917-21. [Crossref] [PubMed]
- Zhang N, Gao Y, Huang Z, et al. PARP inhibitor plus radiotherapy reshapes an inflamed tumor microenvironment that sensitizes small cell lung cancer to the anti-PD-1 immunotherapy. Cancer Lett 2022;545:215852. [Crossref] [PubMed]
- Sen T, Rodriguez BL, Chen L, et al. Targeting DNA Damage Response Promotes Antitumor Immunity through STING-Mediated T-cell Activation in Small Cell Lung Cancer. Cancer Discov 2019;9:646-61. [Crossref] [PubMed]


