
Dissecting the genetic variations associated with response to first-line chemotherapy in patients with small cell lung cancer: a retrospective cohort study
Highlight box
Key findings
• An increased epidermal growth factor receptor (EGFR) gene copy number may be a positive biomarker for small cell lung cancer (SCLC) patients treated with first-line standard chemotherapy.
What is known and what is new?
• EGFR mutations are found in a small number of SCLC patients, who may be sensitive to EGFR-tyrosine kinase inhibitors. There are few studies on the impact of prognosis in patients with EGFR copy number amplification.
• We compared the genetic biology of SCLC patients in partial response and progressive disease groups receiving first-line standard chemotherapy and found that EGFR amplification may be associated with treatment effects.
What is the implication, and what should change now?
• EGFR amplification may be a useful biomarker for chemotherapy-responsive patient selection, and further large-scale research should be conducted.
Introduction
Lung cancer as the second most frequently diagnosed malignancy (1) is classified into non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC) in terms of the morphology of lung cancer cells (2). SCLC is an aggressive epithelial tumor showing a high susceptibility to early development and accounts for approximately 14% of all lung cancer cases (3). About two-thirds of SCLC patients also have metastasis (4). Currently, there is limited treatment options available for SCLC, suggesting that SCLC is one of the most fatal malignancies (5).
Regardless of the age, gender, or clinical stage of SCLC patients, chemotherapy is usually an essential part of treatment (6). In recent decades, etoposide and platinum-doublet chemotherapy has been applied as a standard therapy for SCLC, and combination chemotherapy with other agents, including irinotecan (Camptosar), carboplatin (Paraplatin), and DNA topoisomerase inhibitor, have been considered as standard second-line treatment (7-10). To ameliorate adverse reactions and drug resistance to chemotherapy alone, alternative therapeutic options such as radiotherapy, surgery, and/or immunotherapy are often introduced (4,11-14). Combined used of radiotherapy and chemotherapy could improve local control of patients with limited-stage SCLC and their overall survival. The most commonly used initial chemotherapy regimen for those with extensive-stage SCLC is etoposide/cisplatin (EP). However, there is no chemotherapy combinations showing superior efficacy (15).
Because of recent advances in high-resolution detection technology, a new understanding of the genetic biology of SCLC has led to the development of more selective and targeted therapies, the most promising of which is that the genetic variability in individual patients may predict drug response and therapeutic efficacy or susceptibility to adverse drug reactions (16). At common RNA levels, an upregulated miR-27a expression after chemotherapy was seen in partial response (PR) patients than in those who exhibited no response (NR), and further survival analysis indicated that patients with reduced miR-27a levels displayed inferior outcomes than those with raised miR-27a levels (17). Furthermore, in epidermal growth factor receptor (EGFR)-mutant NSCLC patients, EGFR-tyrosine kinase inhibitors (TKIs) were adopted for considerable therapeutic effects (18). Genetic variation was also related to response to dutasteride for male undergoing androgenetic alopecia (19) as well as long-term therapeutic response in bipolar depression (20). What’s more, the combination of genomic variation with other immunotherapy related indicators has been thought to be meaningful for precise immunotherapy decisions for advanced lung squamous cell carcinoma (21). All these findings highlight the importance of genetic variation in drug treatment. Nonetheless, similar studies on SCLC are rare (22,23). A few consistent associations have been reported for some individual susceptibility genes, but no general recommendations have been formulated to date (24-26).
Herein, this retrospective study aimed to investigate the clinical characteristics of genetic variations detected by single-nucleotide polymorphism (SNP) sequencing in tissue samples collected from SCLC patients prior to first-line chemotherapy. Then, the genotyping data of the patients were comprehensively analyzed between the progressive disease (PD) and PR groups. We present this article in accordance with the TRIPOD reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1772/rc).
Methods
Patients and data collection
A total of 24 SCLC patients who underwent first-line standard chemotherapy (EP regimen) from October 2009 to February 2012 were enrolled in this retrospective study. For enrolled patients, they were regularly followed every 3 months with computed tomography (CT) scan until recurrences determined by CT scan results. They were allocated into a PD group (n=10) or a PR group (n=14) based on the curative effect of chemotherapy assessed by CT scan results. PD was defined as the appearance of new lesions or a >25% increase in the size of lesions, while PR was defined as a >50% reduction in the size of the lesions. The inclusion criteria were as follows: (I) patients diagnosed as SCLC by pathology; (II) patients treated with first-line EP chemotherapy; (III) patients who received long-term follow-up and signed informed consent. Patients were excluded based on the following criteria: (I) history of other malignancies; (II) history of myocardial infarction, unstable angina pectoris, stroke, or uncontrollable arrhythmias; (III) pregnant or lactating patients; (IV) history of mental illness; (V) poor compliance. The clinical characteristics of SCLC patients are summarized in Table 1.
Table 1
| Clinical features | EGFR gene copy number | P value | |
|---|---|---|---|
| Gain (n=8) | Non-gain (n=16) | ||
| Gender, n (%) | 0.718 | ||
| Female | 1 (12.5) | 0 | |
| Male | 7 (87.5) | 16 (100.0) | |
| Age | 0.95 | ||
| Mean (SD) | 63.3 (5.63) | 63.1 (8.74) | |
| Median [Min, Max] | 62.5 [57.0, 70.0] | 62.5 [48.0, 76.0] | |
| Stage, n (%) | >0.99 | ||
| IIb–IIIa | 2 (25.0) | 3 (18.8) | |
| IIIb–IV | 6 (75.0) | 13 (81.3) | |
| Limited or extensive stage, n (%) | 0.884 | ||
| Extensive stage | 4 (50.0) | 10 (62.5) | |
| Limited stage | 4 (50.0) | 6 (37.5) | |
EGFR, epidermal growth factor receptor; SD, standard deviation.
The sample size of the trial was determined by the analysis of overall survival. We calculated that 26 deaths in the chemotherapy treated SCLC population would be needed to provide 90% power at a two-sided significance level of 0.05 to detect a significance between treatment-resistance and treatment-sensitive group.
Tumor specimens were acquired by surgery (>2% of total tissue mass and >150 cells). Diagnosis of SCLC was confirmed by pathologists using Formalin-fixed and paraffin-embedded tissues. Tumor-node-metastasis (TNM) staging system of International Association for the Study of Lung Cancer (version 7) was used to determine the clinical staging. Chi-squared test or Fisher’s exact test were analyzed categorical variables for baseline comparability.
The study was approved by the Ethics committee of the First Affiliated Hospital of Guangzhou Medical University (No. 202015). Written informed consent was signed by all the patients. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
DNA extraction
Tissue samples were collected from SCLC patients prior to first-line chemotherapy and embedded in paraffin (5–10 µm thick for each section). Pathological examination was performed to calculate the quantity of tumor content, which was required to be no less than 10%. Then, NovaSeq S-Prime Reagent Kits (Illumina, California, USA) was employed to extract DNA from the paraffin-embedded tissue blocks. DNA concentration and quality was assessed using Qubit dsDNA HS (High Sensitivity) Assay Kit (Thermo Fisher, Waltham, USA) and Agilent 2100 BioAnalyzer (Agilent, Palo Alto, USA), respectively.
Library preparation and next-generation sequencing (NGS)
A total of 74.5 ng DNA per tissue sample was isolated to construct the DNA libraries. Firstly, genomic DNA was sheared into 160–200 bp small fragments by the Covaris LE220 (Covaris, New Jersey, USA). Subsequently, a KAPA Hyper Preparation Kit (Kapa Biosystems, Boston, USA) was used to construct a fragmented DNA library. The libraries were quantified with an AccuGreen High Sensitivity dsDNA Quantitation Kit (Biotium, California, USA), and their size was determined by an Agilent Bioanalyzer 2100 (Agilent). The DNA library was captured by a designed panel of 1,406 cancer-related genes (Genecast Biotechnology Co. Ltd., Beijing, China) that are frequently mutated in common solid tumors and span a 2.4 Mb region in the human genome. Finally, the captured library was subjected to NovaSeq 6000 (Illumina) sequencing, which produced paired-end sequencing with the length of each end as 150 bp.
Sequencing data analysis
After obtaining the NGS sequencing and confirming quality control, the genomic loci were examined and mapped to the human genome reference (HG19) with a Burrows-Wheeler Aligner (BWA, v0.7.17) (27). Subsequently, the differences in single nucleotide variations (SNVs) and deletion (indels) were determined using VarScan2 (RRID: SCR_006849) and the algorithm independently developed by Genecast Biotechnology Company. SNV is mainly based on the pileup of sequencing reads obtained, and the mutation type and frequency can be inferred according to the number of different supporting bases in the same genome location and sequencing quality. To determine the presence of copy number variations (CNVs), CONTRA software (version 2.0.8) was used to call copy number values from the tumor tissue-derived DNA of patients by sequencing depth and comparison with a healthy population baseline, with a copy number threshold of 3 for CNV gain and 1.2 for CNV loss. Copy number instability (CNI) was defined as the sum of the fluctuation levels of all gene copy numbers in the panel relative to 30 normal people randomly selected from the normal population database of Genecast Biotechnology Company. For CNI calculation, the copy numbers were called after mapping using the BWA tool. After applying proprietary algorithms for tumor DNA sequencing to correct for GC-content and mappability, the read counts were converted into log2 ratios and Z-values based on Gaussian transformations compared with 30 normal people. Target areas with a Z-score >95th percentile and twice the absolute standard deviation of the normal control group were retained, and CNI was the sum of these Z-scores.
Calculation of tumor mutational burden (TMB)
TMB refers to the total number of somatic missense mutations in a baseline tumor sample. TMB was calculated based on the number of somatic nonsynonymous SNVs (depth >100× and allele frequency ≥0.05) detected on NGS (interrogating Mb of the genome), followed by extrapolation to the whole exome using the validated algorithm. Alterations that were likely or known to be bona fide oncogenic drivers were excluded. TMB was calculated in mutations per Mb.
Statistical analyses
SPSS version 20.00 was used for the statistical analysis. Fisher’s exact test, nonparametric log-rank test, and Wilcoxon rank-sum test were used to determine the differences in somatic mutations, CNV, TMB, and CNI between the PD and PR groups. The comparison of overall survival between the two groups was analyzed by the Mann-Whitney U test. A statistically significant difference was defined when P value <0.05.
Results
Clinical characteristics of SCLC patients
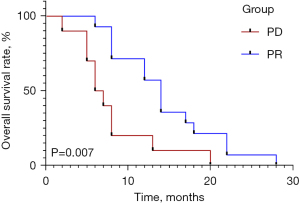
The median age of patients in the PD and PR groups was 62 and 64.5 years, respectively (Table 1). Sex, age, clinical stage, and limited or extensive stage showed no significant differences (all P>0.05) between the PD and PR groups. PD patients showed significantly lower median overall survival than those with PR (6.5 vs. 14.0 months, respectively, P=0.007) (Figure 1).
CNVs in SCLC tissue DNA
As seen in Figure 2A, the incidence of CNV in the PR group was higher than that in the PD group, and there were nine common genes with CNVs in both groups, including FDPS, MAP2K2, PSMA8, MALAT1, DDX27, GSTT1, SDHA, SMAD2, and VEGFA. Compared with the PD group, increased gene copy numbers of EGFR and SQX2-OT were only observed in the PR group (P=0.006 and P=0.053, respectively). Additionally, 21% of patients had an increased SQX2-OT gene copy number gain, relatively close to the 27% detected in our previous study (28). Except for EGFR, CNVs in other genes showed no obvious difference between the two groups using Fisher’s exact test (all P>0.05). Additionally, we also analyzed the relationship between EGFR gene mutation and clinicopathological features of SCLC. Surprisingly, we found that EGFR gene mutation had no obvious connections with clinicopathological features in SCLC patients (Table 2). This observation indicated that EGFR gene mutation may be an independent indicator for SCLC patients.
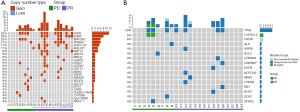
Table 2
| Characteristics | Progressive disease group (n=10) | Partial remission group (n=14) | P value |
|---|---|---|---|
| Male | 10 (100.0) | 13 (92.9) | >0.99 |
| Age (years) | 62.00 [48.00, 76.00] | 64.50 [49.00, 70.00] | 0.827 |
| Clinical stage | 0.441 | ||
| IIb–IIIa | 2 (20.0) | 3 (21.4) | |
| IIIb–IV | 8 (80.0) | 11 (78.6) | |
| Limited or extensive stage | 0.4212 | ||
| Limited stage | 3 (30.0) | 7 (50.0) | |
| Extensive stage | 7 (70.0) | 7 (50.0) | |
| Overall survival (months) | 6.50 [5.00, 9.25] | 14.00 [8.00, 19.00] | 0.007 |
Data are presented as median [interquartile range] or n (%). EGFR, epidermal growth factor receptor; SCLC, small cell lung cancer.
Comparison of somatic mutations between the PD and PR groups
The related genes and SNP sites of chemotherapy drugs for SCLC recommended by the National Comprehensive Cancer Network (NCCN) guidelines in PharmGKB (https://www.pharmgkb.org/) (29) were included in our study. A tumor tissue-derived DNA single nucleotide mutation map before first-line EP treatment showed that the TP53 gene mutation was detected in four cases in the PD group and three cases in the PR group, with no significant difference (P=0.393) (Figure 2B). In addition, genes of nonsynonymous SNV (BCL2, SOX2, and ZFHX3), non-frameshift deletion (FGFR1), and stopgain (CYP2C19) were found in the PD group, while genes of nonsynonymous SNV (ASPM, CDKN2A, CREBBP, EGFR, NOTCH2, NRAS, PTPRB, RB1, and ROS1) and non-frameshift deletion (FGFR1) were identified in the PR group (all P>0.05).
Analysis of chemotherapy-related gene loci in tumor tissue-derived DNA
To analyze the differences in the genotype of chemotherapy-related gene loci in tumor tissue-derived DNA between the PD and PR groups, we selected six genotypes related to first-line treatment efficacy (DYNC2H1_rs716274, ERCC1_rs11615, XRCC1_rs25487, GSTP1_rs1695, XPC_rs2228001, and SLIT1_rs2784917). The correlation between chemotherapy efficacy and the six genotypes was analyzed with the log-rank test. Results showed no significant difference in the genotype of chemotherapy-related gene loci between the two groups (all P>0.05) (Figure 3).

Analysis of TMB and CNI
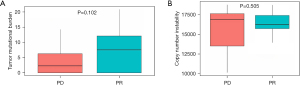
As shown in Figure 4A, the TMB of tumor tissues from SCLC patients in the PR group was higher than in the PD group, but there was no significant difference (P=0.102). Likewise, no significant difference was observed in the CNI of tissue DNA between the two groups (P=0.505) (Figure 4B).
Discussion
SCLC is a metastatic malignancy with high recurrence and poor prognosis. Of the 24 patients recruited in this study, more than half had extensive-stage SCLC, suggesting that the cancer had spread to other organs, such as the other lung, bone, brain, or bone marrow. This retrospective analysis aimed to establish a landscape of genetic variations in Chinese SCLC patients receiving standard first-line chemotherapy and to correlate these variation profiles with clinical outcomes, including remission status and survival time. Our results demonstrated significant differences in the CNVs of increased EGFR gene copy numbers and overall survival time between patients who achieved PR after first-line chemotherapy and patients who developed PD.
In China, the commonly mutated genes in SCLC include TP53, RB1, LRP1B, and PTEN (30). In addition, a study by Xu et al. found that RB1 mutations were found in 83.6% of patients with SCLC in China (31). In our study, NGS with a designed panel was performed on tumor tissue-derived DNA to identify the gene mutations related to SCLC. We found that the commonly mutated genes in SCLC patients in the PD and PR groups were nonsynonymous SNV of TP53 and indels of FGFR1, with no significant difference. Yokouchi et al. (32) have pointed out that the nonsynonymous somatic TP53 mutation in SCLC patients is an independent factor for prolonged relapse-free survival rather than overall survival. Nevertheless, no significant differences in SNV of TP53 and indels of FGFR1 were found between the two groups.
It has been reported that widespread somatic CNVs included MYC amplification, copy number gains of SOX2 at 3q26, CCNE1 at 19q12, FGF10 at 5p13, and SOX2 at 3q26, and copy number losses of FHIT at 3p14, PTEN at 10q23, RASSF1 at 3p21.3, TP53 at 17p13, and RB1 at 13q14.2 (28). Our data showed that the common genes involved in CNVs included FDPS, EGFR, MAP2K2, MALAT1, DDX27, GSTT1, SDHA, SMAD2, and VEGFA, which was partially different from our previous study. However, we repeatedly checked the data of the detection results and found no RB1 alteration or MYC amplification, which could be explained by the small sample size. Further group comparison showed a statistically significant difference in the increased EGFR gene copy number between the PD and PR groups (P<0.05).
EGFR is a member of the erbB family of tyrosine kinase receptors, and the EGFR gene coding the receptor is localized at chromosome 7 (33). The normal physiological function of EGFR is to regulate epithelial tissue development and homeostasis. However, under pathological environment, such as lung, breast cancer as well as glioblastoma, it was demonstrated to be a driver of tumorigenesis (34). As research deepens, abnormally activated EGFR in tumor mainly accounts for amplification and point mutations at the genomic locus, which lead to unfavorable survival (35,36) and tolerance to various chemotherapy drugs (37). These findings drive the predicted value of EGFR gene copy number. Therefore, given that EGFR mutations are detected in up to 50% of NSCLC, EGFR has also become a critical target in NSCLC treatment (38). Higaki et al. further observed that patients with EGFR gene copy number gain but not amplification, including those exhibiting polysomy, also exhibited poorer prognosis than gene copy number non-gain patients. This study indicated that the higher mutation status may result in greater drug tolerance (36). In contrast, EGFR gene mutations are rare in SCLC, accounting for only 2.6–7.1% of SCLC patients in China (39). In 2006, Okamoto et al. were the first to report an EGFR mutation (heterozygous in-frame 15-base pair deletion) in a gefitinib-responsive SCLC patient (40). Two years later, Tatematsu et al. examined the EGFR gene copy number in five SCLC patients with EGFR mutations and found gene amplification in four cases (41). Since then, some SCLC cases with EGFR mutations have been reported successively, showing that EGFR mutations are sensitive to EGFR-TKIs and may suggest a positive prognostic efficacy (42-44). Conversely, one prior report has clarified that EGFR is low expressed in SCLC, suggesting that EGFR-TKIs are ineffective against SCLC even when EGFR is mutated (41).
Despite its rapid growth and early metastasis, SCLC is more chemosensitive and radiosensitive than other lung cancers. Chemotherapy containing EP has been the standard therapy for extensive-stage SCLC for decades (6). Kim et al. have clarified patients treated with EP regimen showed an average rate of response of 60% to 80%, with a median overall survival of 8 to 10 months (45). This study demonstrated a noticeably better survival of patients in the PR group than in the PD group (14 vs. 6.5 months). As mentioned, PR was achieved in eight SCLC patients with an increased EGFR gene copy number who were treated with the EP regimen. Taken together, the longer overall survival in the PR group indicated a positive therapeutic effect of standard first-line chemotherapy in Chinese SCLC patients, which might be related to the increased EGFR gene copy number.
To determine the differences in the genotype of chemotherapy-related gene loci between the PD and PR groups, six genotypes related to first-line treatment efficacy, including DYNC2H1_rs716274, ERCC1_rs11615, XRCC1_rs25487, GSTP1_rs1695, XPC_rs2228001, and SLIT1_rs2784917 were selected. Results showed no significant difference in the genotypes of chemotherapy-related gene loci between the two groups. However, one report that analyzed the association and multiple interactions of five XRCC1 polymorphic variants in regulating lung cancer risk in a North Indian population described a positive correlation between XRCC1 Gln632Gln and lung cancer, whereas Arg399Gln, XRCC1, and Arg194Trp had no protective effect (46). In addition, in the present study, the TMB between the PD and PR groups was not significantly different. Likewise, no significantly different CNI value in the tumor tissue-derived DNA of SCLC patients was noticed between the two groups.
Conclusions
In summary, SCLC patients in the PR group showed higher overall survival after first-line chemotherapy (EP regimen) treatment, which may be associated with the increased EGFR gene copy number. This study has some limitations, such as the lack of immunohistochemical analysis and constitutional genetic information for the SCLC patients, as well as the limited sample size. Tissue DNA from only 24 cases may have caused less tumor cells to be tested, which may be the reason for the fewer gene mutations detected. In addition, the lack of a control group may have led to an inability to accurately filter out germ-line mutations, which may have affected the analysis results. Patients in this study ranged from stages II to IV, but the molecular typing of patients at different stages differs, which requires further study based on a fixed stage of SCLC patients in the future.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1772/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1772/dss
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1772/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1772/coif). H.Z. is from Genecast Biotechnology Co. Ltd., Wuxi, Jiangsu, China. J.Z. is from HaploX Biotechnology, Shenzhen, Guangdong, China. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Ethics committee of the First Affiliated Hospital of Guangzhou Medical University (No. 202015). Written informed consent was signed by all the patients. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Soo RA, Stone ECA, Cummings KM, et al. Scientific Advances in Thoracic Oncology 2016. J Thorac Oncol 2017;12:1183-209. [Crossref] [PubMed]
- Li Q, Wang R, Yang Z, et al. Molecular profiling of human non-small cell lung cancer by single-cell RNA-seq. Genome Med 2022;14:87. [Crossref] [PubMed]
- Waqar SN, Morgensztern D. Treatment advances in small cell lung cancer (SCLC). Pharmacol Ther 2017;180:16-23. [Crossref] [PubMed]
- Koinis F, Kotsakis A, Georgoulias V. Small cell lung cancer (SCLC): no treatment advances in recent years. Transl Lung Cancer Res 2016;5:39-50. [Crossref] [PubMed]
- Dingemans AC, Früh M, Ardizzoni A, et al. Small-cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up Ann Oncol 2021;32:839-53. [Crossref] [PubMed]
- Lu H, Jiang Z. Advances in antibody therapeutics targeting small-cell lung cancer. Adv Clin Exp Med 2018;27:1317-23. [Crossref] [PubMed]
- Han D, Wang G, Sun L, et al. Comparison of irinotecan/platinum versus etoposide/platinum chemotherapy for extensive-stage small cell lung cancer: A meta-analysis. Eur J Cancer Care (Engl) 2017;26: [Crossref] [PubMed]
- Chen Y, Chen L, Zhong D. Comparing the adverse effects of platinum in combination with etoposide or irinotecan in previously untreated small-cell lung cancer patients with extensive disease: A network meta-analyses. Thorac Cancer 2017;8:170-80. [Crossref] [PubMed]
- Shi Y, Hu Y, Hu X, et al. Cisplatin combined with irinotecan or etoposide for untreated extensive-stage small cell lung cancer: A multicenter randomized controlled clinical trial. Thorac Cancer 2015;6:785-91. [Crossref] [PubMed]
- Kim YH, Mishima M. Second-line chemotherapy for small-cell lung cancer (SCLC). Cancer Treat Rev 2011;37:143-50. [Crossref] [PubMed]
- Wolf M, Tebbe S, Fink T. First-line chemotherapy in metastatic small-cell lung cancer (SCLC). Lung Cancer 2004;45:S223-34. [Crossref] [PubMed]
- Liu SV, Reck M, Mansfield AS, et al. Updated Overall Survival and PD-L1 Subgroup Analysis of Patients With Extensive-Stage Small-Cell Lung Cancer Treated With Atezolizumab, Carboplatin, and Etoposide (IMpower133). J Clin Oncol 2021;39:619-30. [Crossref] [PubMed]
- Sathiyapalan A, Febbraro M, Pond GR, et al. Chemo-Immunotherapy in First Line Extensive Stage Small Cell Lung Cancer (ES-SCLC): A Systematic Review and Meta-Analysis. Curr Oncol 2022;29:9046-65. [Crossref] [PubMed]
- Horiike A, Saijo N. Treatment of Small Cell Lung Cancer. Cancer Res Treat 2003;35:177-80. [Crossref] [PubMed]
- Zhang Y, Wang DC, Shi L, et al. Genome analyses identify the genetic modification of lung cancer subtypes. Semin Cancer Biol 2017;42:20-30. [Crossref] [PubMed]
- Xie E, Lin M, Sun Z, et al. Serum miR-27a is a biomarker for the prognosis of non-small cell lung cancer patients receiving chemotherapy. Transl Cancer Res 2021;10:3458-69. [Crossref] [PubMed]
- Han R, Jia Y, Li X, et al. Concurrent use of metformin enhances the efficacy of EGFR-TKIs in patients with advanced EGFR-mutant non-small cell lung cancer-an option for overcoming EGFR-TKI resistance. Transl Lung Cancer Res 2021;10:1277-91. [Crossref] [PubMed]
- Rhie A, Son HY, Kwak SJ, et al. Genetic variations associated with response to dutasteride in the treatment of male subjects with androgenetic alopecia. PLoS One 2019;14:e0222533. [Crossref] [PubMed]
- Anmella G, Vilches S, Espadaler-Mazo J, et al. Genetic Variations Associated with Long-Term Treatment Response in Bipolar Depression. Genes (Basel) 2021;12:1259. [Crossref] [PubMed]
- Xu Y, Li H, Huang Z, et al. Predictive values of genomic variation, tumor mutational burden, and PD-L1 expression in advanced lung squamous cell carcinoma treated with immunotherapy. Transl Lung Cancer Res 2020;9:2367-79. [Crossref] [PubMed]
- Zhang Y, Zhang L, Li R, et al. Genetic variations in cancer-related significantly mutated genes and lung cancer susceptibility. Ann Oncol 2017;28:1625-30. [Crossref] [PubMed]
- Pu X, Hildebrandt MA, Lu C, et al. Inflammation-related genetic variations and survival in patients with advanced non-small cell lung cancer receiving first-line chemotherapy. Clin Pharmacol Ther 2014;96:360-9. [Crossref] [PubMed]
- Han S, Gao F, Yang W, et al. Identification of an SCLC susceptibility rs7963551 genetic polymorphism in a previously GWAS-identified 12p13.33 RAD52 lung cancer risk locus in the Chinese population. Int J Clin Exp Med 2015;8:16528-35.
- Yang X, Gao F, Ma F, et al. Association of the functional BCL-2 rs2279115 genetic variant and small cell lung cancer. Tumour Biol 2016;37:1693-8. [Crossref] [PubMed]
- Hellmann MD, Callahan MK, Awad MM, et al. Tumor Mutational Burden and Efficacy of Nivolumab Monotherapy and in Combination with Ipilimumab in Small-Cell Lung Cancer. Cancer Cell 2018;33:853-861.e4. [Crossref] [PubMed]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26:589-95. [Crossref] [PubMed]
- Rudin CM, Durinck S, Stawiski EW, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 2012;44:1111-6. [Crossref] [PubMed]
- Whirl-Carrillo M, Huddart R, Gong L, et al. An Evidence-Based Framework for Evaluating Pharmacogenomics Knowledge for Personalized Medicine. Clin Pharmacol Ther 2021;110:563-72. [Crossref] [PubMed]
- Du M, Thompson J, Fisher H, et al. Genomic alterations of plasma cell-free DNAs in small cell lung cancer and their clinical relevance. Lung Cancer 2018;120:113-21. [Crossref] [PubMed]
- Xu G, Zheng J, Wang S, et al. Landscape of RB1 alterations in 22,432 Chinese solid tumor patients. Ann Transl Med 2022;10:885. [Crossref] [PubMed]
- Yokouchi H, Nishihara H, Harada T, et al. Detection of somatic TP53 mutation in surgically resected small-cell lung cancer by targeted exome sequencing: association with longer relapse-free survival. Heliyon 2020;6:e04439. [Crossref] [PubMed]
- Chinchilla-Tábora LM, Sayagués JM, González-Morais I, et al. Prognostic Impact of EGFR Amplification and Visceral Pleural Invasion in Early Stage Pulmonary Squamous Cell Carcinomas Patients after Surgical Resection of Primary Tumor. Cancers (Basel) 2022;14:2174. [Crossref] [PubMed]
- Sigismund S, Avanzato D, Lanzetti L. Emerging functions of the EGFR in cancer. Mol Oncol 2018;12:3-20. [Crossref] [PubMed]
- Chang N, Duan J, Wang L, et al. Patients with advanced non-small cell lung cancer with EGFR mutations in addition to complex mutations treated with osimertinib have a poor clinical outcome: A real-world data analysis. Oncol Lett 2020;20:2266-72. [Crossref] [PubMed]
- Higaki E, Kuwata T, Nagatsuma AK, et al. Gene copy number gain of EGFR is a poor prognostic biomarker in gastric cancer: evaluation of 855 patients with bright-field dual in situ hybridization (DISH) method. Gastric Cancer 2016;19:63-73. [Crossref] [PubMed]
- Blaquier JB, Ortiz-Cuaran S, Ricciuti B, et al. Tackling Osimertinib Resistance in EGFR-Mutant Non-Small Cell Lung Cancer. Clin Cancer Res 2023;29:3579-91. [Crossref] [PubMed]
- Du X, Yang B, An Q, et al. Acquired resistance to third-generation EGFR-TKIs and emerging next-generation EGFR inhibitors. Innovation (Camb) 2021;2:100103. [Crossref] [PubMed]
- Hu J, Wang Y, Zhang Y, et al. Comprehensive genomic profiling of small cell lung cancer in Chinese patients and the implications for therapeutic potential. Cancer Med 2019;8:4338-47. [Crossref] [PubMed]
- Okamoto I, Araki J, Suto R, et al. EGFR mutation in gefitinib-responsive small-cell lung cancer. Ann Oncol 2006;17:1028-9. [Crossref] [PubMed]
- Tatematsu A, Shimizu J, Murakami Y, et al. Epidermal growth factor receptor mutations in small cell lung cancer. Clin Cancer Res 2008;14:6092-6. [Crossref] [PubMed]
- Bordi P, Tiseo M, Barbieri F, et al. Gene mutations in small-cell lung cancer (SCLC): results of a panel of 6 genes in a cohort of Italian patients. Lung Cancer 2014;86:324-8. [Crossref] [PubMed]
- Siegele BJ, Shilo K, Chao BH, et al. Epidermal growth factor receptor (EGFR) mutations in small cell lung cancers: Two cases and a review of the literature. Lung Cancer 2016;95:65-72. [Crossref] [PubMed]
- Cardona AF, Rojas L, Zatarain-Barrón ZL, et al. Multigene Mutation Profiling and Clinical Characteristics of Small-Cell Lung Cancer in Never-Smokers vs. Heavy Smokers (Geno1.3-CLICaP). Front Oncol 2019;9:254. [Crossref] [PubMed]
- Kim DW, Kim HG, Kim JH, et al. Randomized Phase III Trial of Irinotecan Plus Cisplatin versus Etoposide Plus Cisplatin in Chemotherapy-Naïve Korean Patients with Extensive-Disease Small Cell Lung Cancer. Cancer Res Treat 2019;51:119-27. [Crossref] [PubMed]
- Singh A, Singh N, Behera D, et al. Association and multiple interaction analysis among five XRCC1 polymorphic variants in modulating lung cancer risk in North Indian population. DNA Repair (Amst) 2016;47:30-41. [Crossref] [PubMed]
(English Language Editor: D. Fitzgerald)


