
Transcription factor-target gene regulatory network analysis in human lung adenocarcinoma
Highlight box
Key findings
• This study constructed a regulatory network of key transcription factors (TFs) and target genes involved in lung adenocarcinoma (LUAD). Here, two potential key TFs involved in the regulation of LUAD (GRHL3 and TAL1) were identified, along with five alternative splicing events (XMIR, XIR, XAE, MIR, and IR) that exhibited abnormal occurrence in LUAD.
What is known and what is new?
• The regulatory TF-target gene networks involved in the development of LUAD is complex and not yet fully understood.
• Here, we propose the existence of a GRHL3-CDH15-Wnt-β-catenin pro-oncogenic signaling axis and a TAL1-ADAMTS1-vascular antioncogenic signaling axis.
What is the implication, and what should change now?
• The result indicated a novel TF-target gene regulatory axis related to tumorigenesis and provided potential therapeutic targets and mechanisms for LUAD. Although further experimental validation is warranted, this study provides potential targets and clues for the diagnosis and treatment of LUAD in the future.
Introduction
Lung cancer is a common malignant tumor that originates from malignant cells in the lung tissue and is one of the leading causes of death worldwide (1,2). Lung adenocarcinoma (LUAD) is one of the most common subtypes of lung cancer, occurring in the epithelial cells of the LUAD and accounting for approximately 40% of all lung cancer cases. The etiology and pathogenesis of lung cancer are complex and closely related to genetic, environmental, and lifestyle factors. The pathogenesis of lung cancer involves multiple molecular pathways and signaling transduction pathways. Studies have shown that activation of proliferative signaling pathways, inactivation of tumor-suppressor genes and DNA repair genes, and epigenetic modifications can lead to the development of lung cancer (3). In addition, inflammation, immune cells, and cytokines in the tumor microenvironment are also closely associated with the occurrence of lung cancer (3,4). Currently, the treatment modalities for lung cancer include surgical resection, radiotherapy, chemotherapy, genetic testing, immunotherapy, and targeted therapy (5-7). Although these treatment modalities have played important roles in lung cancer treatment, there are still challenges and bottlenecks, including tumor drug resistance, difficulties in early diagnosis, individual differences, and genetic heterogeneity.
Transcription factors (TFs) are a class of proteins widely present in organisms and play a regulatory role in gene expression. In the progress and development of LUAD, TFs participate in processes such as cell proliferation, apoptosis, invasion, and metastasis through their regulatory functions. Previous studies have shown that many TFs exhibit specific expression patterns in LUAD. For example, the expression level of FOXP3 is often low in LUAD tissues and is associated with prognosis (8). Other TFs such as p63 and TTF-1 can be used for the diagnosis and differentiation of different types of LUAD (9). The MYC family of TFs is critically involved in the occurrence and development of LUAD (10). Overexpression of c-Myc can promote the proliferation and survival of LUAD cells and enhance their invasive and metastatic abilities. N-Myc can regulate the expression of various LUAD-related genes, such as BCL2, MCL1, and BMI1, thereby closely influencing the occurrence and development of LUAD. Inactivation of TP53 can lead to abnormal cell cycle and apoptosis inhibition in LUAD, thus promoting malignant transformation and progression. TP53 also regulates the expression of multiple genes related to LUAD, such as MDM2, p21, and BAX, also contributing to the occurrence and development of LUAD (11). Considering the critical role of TFs in the progress of LUAD, targeting TFs has become an important direction in the treatment of LUAD. Currently, many targeted therapy drugs against c-Myc are under development, which inhibit the growth and metastasis of LUAD by suppressing the expression or activity of c-Myc (10). In addition, due to the significant role of TP53 in LUAD, several treatment strategies targeting TP53 are also under investigation. For example, synthetic peptides targeting the core region of TP53 can bind to MDM2, inhibiting MDM2-mediated degradation of TP53 and restoring its function, thus providing a therapeutic approach for LUAD (12).
High-throughput sequencing technology, through comprehensive analysis of all transcripts in LUAD cells or tissues, has assisted in gaining a deeper understanding of the pathogenesis of LUAD and identifying multiple TFs involved in its occurrence and development (4,13,14). A recent transcriptional regulatory network study identified key TFs involved in LUAD development, including SOX10, SPIB, NR4A2, FOXD1, ELF5, HOXA5, KLF5, ESRRA, SREBF1 and REL (14). However, due to the diversity and heterogeneity of LUAD, our knowledge of the TF regulatory network in LUAD remains limited, particularly in terms of downstream target gene regulation networks and signaling pathways. In this study, we continued to use high-throughput sequencing for transcriptomic analysis of normal adjacent tissues and LUAD tissues in humans. We identified differentially expressed TFs in LUAD and discovered several TFs that were significantly upregulated in LUAD tissues, including GRHL3, SIX1, SIX2, SPDEF, and ETV4, while TAL1, EPAS1, SOX17, NR4A1, and EGR3 were significantly downregulated. Subsequently, we constructed a key differential TF-target gene regulatory network by combining differentially expressed genes. Among them, the TFs GRHL3 and TAL1 exhibited the most significant differential expression in LUAD tissues, with GRHL3 being newly discovered as being upregulated in LUAD tissues. Therefore, we further predicted target genes and conducted functional analysis of these potential target genes. The expression of GRHL3 and TAL1 were positively correlated with the survival rate of patients with LUAD. Additionally, to further analyze the transcriptional regulatory mechanisms during the development of LUAD, we examined selective splicing events, an important posttranscriptional regulatory mechanism, and found that intron retention (IR), approximate IR (XIR), multi-IR (MIR), approximate MIR (XMIR), and approximate alternative exon ends (XAE) showed abnormally increased frequencies in LUAD tissues. In summary, for the first time, we constructed a differential TF-target gene regulatory network in LUAD tissues and proposed a potential GRHL3-CDH15-Wnt-β-catenin pro-oncogenic signaling axis and a potential TAL1-ADAMTS1-vascular antioncogenic signaling axis, providing novel mechanistic insights into the targeted TF therapy of LUAD. We present this article in accordance with the STREGA reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1688/rc).
Methods
Human LUAD and paracancerous tissue collection
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Ethics Committee of Binhai County People’s Hospital of Yancheng (No. 2021BHKYLL0023) and informed consent was taken from all the patients. The patients’ clinical information, such as age, gender, tumor node metastasis (TNM) classification, and stage, was basically consistent (Table 1). Fresh LUAD tissues and matched adjacent nontumor tissues were collected from four patients with LUAD who underwent surgical resection at Binhai County People’s Hospital of Yancheng from February 2021 to October 2022. Prior to surgery, none of the patients had received radiotherapy or chemotherapy, had a history of other treatments, or had inflammatory diseases. Tissue specimens collected from LUAD patients were immediately frozen and stored at −196 ℃ in liquid nitrogen for further study.
Table 1
| Patient No. | Age (years) | Gender | Side | T | N | M |
|---|---|---|---|---|---|---|
| 1 | 73 | Male | LL | T1 | N0 | M0 |
| 2 | 71 | Male | RL | T1 | N0 | M0 |
| 3 | 79 | Male | LL | T1 | N0 | M0 |
| 4 | 68 | Male | RL | T1 | N0 | M0 |
LL, left lung; RL, right lung.
RNA sequencing (RNA-seq) and screening of differentially expressed genes
Total RNA from LUAD tissues or matched adjacent nontumor tissues was extracted with Trizol reagent (MilliporeSigma, Burlington, MA, USA) according to the manufacturer’s instructions. Subsequent RNA-seq was conducted by Shanghai OE Biotech Co., Ltd. (Shanghai, China). The differentially expressed genes were screened using fold change, and P values were calculated using the t-test. The threshold set for upregulated and downregulated genes was a fold change ≥2 and a P value ≤0.05.
Enrichment analysis
For enrichment analysis, the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (https://www.genome.jp/kegg/) and Gene Ontology (GO) database (https://geneontology.org/) were used to perform pathway and biological function analyses on these differentially expressed messenger RNAs (mRNAs).
Construction of a differential TF-target gene regulatory network
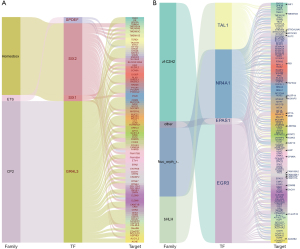
The Gene Transcription Regulation Database (GTRD) (https://gtrd.biouml.org/) was used to predict the interaction between differential TFs and target genes and to construct a differential TF-target gene regulatory network. Based on the prediction results of differential genes and differential TFs (fold change >5), a Sankey diagram of the relationship of the TF family and the TF with the target genes was drawn using OECloud tools (https://cloud.oebiotech.com).
Survival curve analysis
Using the most authoritative website for survival analysis, the Kaplan-Meier plotter database (https://kmplot.com/analysis), we analyzed the impact of the differentially expressed TFs (GRHL3, ETV4, SPDEF, SIX1, SIX2, TAL1) and target gene (CDH15) on the survival rate of patients with LUAD.
Prediction of binding sites of TFs and target genes
To further identify the target gene of GRHL3 and TAL1, the hTFtarget database (http://bioinfo.life.hust.edu.cn/hTFtarget) was used to predict the target genes of these two TFs. The overlap of target genes between the hTFtarget database and the GTRD were selected for further analysis. The JASPAR database (https://jaspar.genereg.net/) was used to predict the binding motifs of GRHL3 (or TAL1) as well as the binding sites between GRHL3 (or TAL1) and CDH15 (or ADAMTS1).
Alternative splicing analysis
The alternatively splicing analysis of differentially regulated transcripts isoforms or exons was performed using ASprofile software. The ASprofile software categorizes alternative splicing events into 12 different types: alternative 5’ first exon (TSS), alternative 3’ last exon (TTS), skipped exon (SKIP), approximate SKIP (XSKIP), multiexon SKIP (MSKIP), approximate MSKIP (XMSKIP), IR, XIR, MIR, XMIR, alternative exon ends (AE), and XAE.
Statistical analysis
In the RNA-seq data, a P value less than 0.05 and a fold change greater than 2 indicated there to be a differential expression of the same gene between the two samples.
Results
Identification of differentially expressed genes in LUAD
To explore the transcriptomic changes during the development of LUAD, we first collected clinical samples of LUAD tissue and adjacent nontumor tissue and then performed RNA-seq to construct libraries for screening differentially expressed genes in LUAD. We obtained results where the original base number for each sample was greater than 7.1 G, the percentage of effective bases was greater than 91%, and the percentage of bases that have a Q-score ≥30 (Q30) was greater than 94%. At least 97% of reads matched the human genome, indicating a high reliability of the sequencing (Table 2). Principal component analysis (PCA) of the sequencing results revealed that the four normal adjacent tissue samples clustered together, indicating a high similarity between normal samples, while one sample from the four LUAD tissues deviated from the other three samples, which may be related to the heterogeneity of lung cancer (Figure 1A). Using a threshold of fold change greater than 2-fold as the cutoff, we identified 1,014 downregulated differentially expressed genes and 681 upregulated differentially expressed genes in LUAD tissue (Figure 1B). Among the top 10 upregulated genes identified, HOXC13, HOXC10, and HOXC11 have been reported to play a role in promoting LUAD, which indicated the accuracy of our sequencing results (8,15,16). We further conducted molecular functional, cellular process, cellular component, and signaling pathway enrichment analyses on these differentially expressed genes. We found that these genes were mainly enriched in molecular functions such as calcium ion binding and Wnt-activated receptor activity; biological processes such as cell adhesion, angiogenesis, and vasculogenesis; cellular components such as plasma membrane and integral component of the plasma membrane (Figure 1C); and signaling pathways such as cell adhesion molecules, vascular smooth muscle contraction, and Wnt signaling pathway (Figure 1D). In conclusion, the abovementioned data indicated that abnormal regulation of cell adhesion, the Wnt signaling pathway, and vascular regulation occurs in LUAD, which is closely related to the development of LUAD.
Table 2
| Sample | Raw bases (G) | Valid bases (%) | Q30 (%) | Mapped reads (%) |
|---|---|---|---|---|
| NM1 | 7.27 | 93.28 | 95.08 | 97.28 |
| NM2 | 7.40 | 92.27 | 95.09 | 97.59 |
| NM3 | 7.20 | 92.27 | 95.05 | 98.19 |
| NM4 | 7.47 | 92.27 | 95.11 | 98.10 |
| LC1 | 7.67 | 92.55 | 95.12 | 97.82 |
| LC2 | 7.59 | 91.81 | 94.95 | 96.92 |
| LC3 | 7.12 | 91.65 | 95.00 | 97.49 |
| LC4 | 7.35 | 90.59 | 95.01 | 97.63 |
RNA-seq, RNA sequencing; Q30, percentage of bases that have a Q-score ≥30; NM, matched adjacent nontumor tissues; LC, LUAD tissues; LUAD, lung adenocarcinoma.
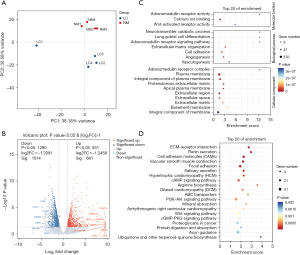
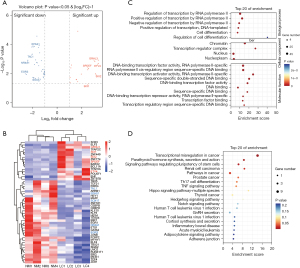
Differentially expressed TFs in LUAD
To further explore the mechanism for the significant transcriptomic dysregulation in LUAD, we focused on TFs and selected a total of 50 differentially expressed TFs from the differentially expressed genes, including 34 downregulated and 16 upregulated TFs (Figure 2A,2B). TFs GRHL3, SIX1, SIX2, SPDEF, and ETV4 were the most significantly upregulated, while TAL1, EPAS1, SOX17, NR4A1, and EGR3 were the most significantly downregulated in LUAD tissue. Among them, SIX1 (17), SIX2 (18), ETV4 (19), EPAS1 (20), and NR4A1 (21) have been shown to be closely associated with the proliferation, invasion, and metastasis abilities of lung cancer, further confirming the accuracy of our identified TFs and suggesting the potential value of the remaining unverified TFs in LUAD. The enrichment analysis results showed that these TFs were mainly enriched in cellular components such as chromatin and nucleus; biological processes such as regulation of transcription by RNA polymerase II and cell differentiation; relevant molecular functions such as DNA-binding TF activity; and signaling pathways such as transcriptional misregulation in cancer (Figure 2C,2D). These data indicate the important role of TFs in the transcriptomic dysregulation of LUAD.
Construction of key differential TF-target gene interaction networks
The functional role of TFs primarily relies on their transcriptional regulation of downstream target gene expression. Although previous studies have revealed the expression and function of some TFs in LUAD, no studies to date have systematically analyzed the target genes regulated by these abnormally expressed TFs in LUAD. To further explore the TF regulatory mechanisms in LUAD, we used GTRD to predict the regulatory networks of the top five upregulated and downregulated differential TFs with their target genes in LUAD based on the differentially expressed genes and TFs (with a fold change threshold >5). In this network, among the top five upregulated and downregulated TFs, one TF in each group was not predicted to have consistent expression with its target genes, while the expression trends of the remaining four TFs were consistent with their corresponding target genes (Figure 3A,3B).
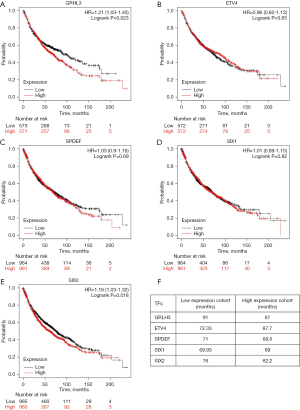
GRHL3 may act as a potential proto-oncogene by transcriptionally regulating CDH15 expression and subsequently regulating Wnt signaling pathway in LUAD
To further understand the functions of these dysregulated TFs in the development of LUAD, we conducted survival analysis using the authoritative Kaplan-Meier plotter database to examine the impact of the top five upregulated differential TFs (GRHL3, ETV4, SPDEF, SIX1, SIX2) on the survival rate of patients with LUAD (Figure 4). We found that the TF GRHL3 had the most significant effect on the survival rate of these patients. Patients with high expression of GRHL3 exhibited a significantly shortened lifespan of up to 24 months compared to those with low expression of GRHL3 (Figure 4A,4F), suggesting that GRHL3 may act as an oncogene and is associated with poor prognosis in LUAD. Among the other 4 TFs, only SIX1 showed a correlation with poor prognosis in LUAD although SIX1, SIX2, and ETV4 have been demonstrated to promote proliferation and migration of non-small LUAD cells (17-19). Patients with a high expression of SIX2 had a lifespan shortened by 14 months compared to those with low expression of SIX2, while SIX1, ETV4, and SPDEF did not have significant associated on the survival rate of patients with LUAD (Figure 4B-4F). Therefore, we next focused on investigating the functions and regulatory mechanisms of GRHL3 in LUAD.
Through enrichment analysis of the predicted target genes of GRHL3 obtained from GTRD, it was found that the target genes of GRHL3 were mainly enriched in the cell adhesion molecules and Wnt signaling pathway (Figure 5A), which are also significantly enriched in the differential genes of LUAD (Figure 1D), suggesting that GRHL3 may play an important role in the transcriptional dysregulation of LUAD. The hTFtarget database contains the most comprehensive information on human TF-target interactions, integrating large-scale human TF-target gene data (22). Therefore, we further predicted the target genes of GRHL3 using the hTFtarget database and performed Venn diagram analysis to identify the intersection between the predicted target genes from GTRD and the predicted target genes from the hTFtarget database, resulting in 22 potential target genes (Figure 5B). Among these 22 potential targets, CDH15 was enriched in the cell adhesion molecules signaling pathway and exhibited significant upregulation in LUAD (Figure 5C). Survival curve analysis revealed that patients with high expression of CDH15 had a significantly shortened lifespan of up to 11 months compared to those with low expression of CDH15 (Figure 5D), further indicating the potential regulatory role of CDH15 in the development of LUAD. By comparing the conserved binding motifs of GRHL3 (23) with the promoter sequence of CDH15, two potential binding sites, 5'-AAGCCGGCT-3' and 5'-GAACAGGTG-3', were identified in the CDH15 promoter (Figure 5E), further suggesting that CDH15 is a potential target gene of GRHL3. CDH15 is an M-cadherin, and cadherins have been found to positively regulate myogenic differentiation induced by the canonical Wnt signaling pathway by regulating the phosphorylation of GSK-3β and β-catenin (24). The abovementioned results indicate that GRHL3 may regulate the Wnt signaling pathway by modulating the expression of the cell adhesion molecule CDH15, thereby promoting the development of LUAD (Figure 5E).
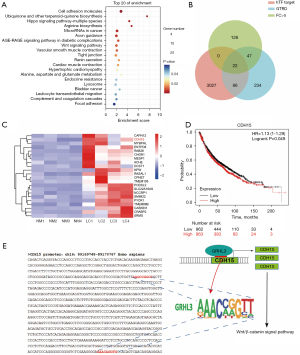
TAL1 may function as a potential tumor-suppressor gene by regulating angiogenesis through transcriptional regulation of ADAMTS1 expression in LUAD
Functional enrichment analysis revealed that differentially expressed genes in LUAD were significantly enriched in the Wnt signaling pathway and vascular regulation pathway (Figure 1C,1D), both of which play crucial roles in the development and progression of LUAD. Through the analysis of significantly upregulated TFs in LUAD, we identified GRHL3 as a potential key TF involved in regulating the Wnt signaling pathway in LUAD (Figure 5). Next, we analyzed the top five downregulated TFs in LUAD (TAL1, EPAS1, SOX17, NR4A1, and EGR3) and found that TAL1 has been reported to be critically involved in angiogenesis and vascular system development (25). TAL1 regulates the expression of VE-cadherin in endothelial cells, contributing to the differentiation and development of endothelial cells and the hematopoietic lineage (26). Therefore, we hypothesized that TAL1 may be a key TF involved in regulating the vascular system in LUAD and subsequently focused on investigating the function and mechanisms of TAL1 in LUAD. Survival curve analysis revealed a positive correlation between TAL1 expression and the survival rate of patients with LUAD. Patients with high TAL1 expression had a significantly extended lifespan of up to 17 months compared to those with low TAL1 expression (Figure 6A), suggesting that TAL1 may function as a tumor suppressor. Through enrichment analysis, we found that the predicted target genes of TAL1 in the GTRD were significantly enriched in the vascular smooth muscle contraction pathway (Figure 6B), which was also significantly enriched in the differentially expressed genes of LUAD (Figure 1D), further highlighting the crucial role of TAL1 in the regulation of the vascular system in LUAD. Furthermore, we used the hTFtarget database to accurately predict the target genes of TAL1 and identified 23 potential target genes through Venn diagram analysis of the predicted target genes from the GTRD and the hTFtarget database (Figure 6C). Among these 23 potential targets, ADAMTS1 was significantly upregulated in LUAD tissues (Figure 6D) and has been reported to be closely associated with angiogenesis and vascular function (27), exhibiting inhibitory effects on some tumors (28). Additionally, through JASPAR database analysis, we identified three potential binding sites of TAL1 in the ADAMTS1 promoter sequence: 5’-TGAACATATCAT-3’, 5’-CAACTTTCTGGT-3’, and 5’-CAGCCCTCTGCT-3’ (Figure 6E). This further suggested that ADAMTS1 as a potential target gene of TAL1. Based on the aforementioned analysis results, we hypothesize that TAL1 may regulate the vascular system of LUAD cells through the transcriptional regulation of ADAMTS1, thereby inhibiting the development and progression of LUAD (Figure 6E).
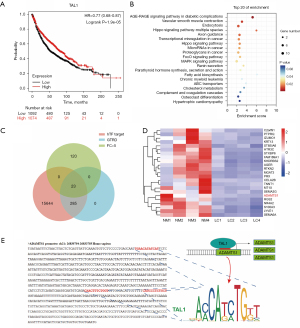
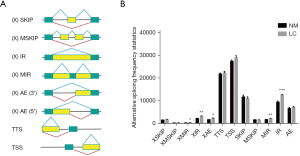
Global identification of dysregulation alternative splicing in LUAD
Alternative splicing is an important posttranscriptional gene regulation mechanism that is closely associated with tumorigenesis. Studies have shown that various abnormal splicing events occur during the development of lung cancer, leading to altered expression and functional changes in tumor-related genes (29-31). For example, selective splicing alterations in genes such as BCL2L1, MDM2, MDM4, NUMB, and MET can affect the apoptosis, proliferation, and cell adhesion of lung cancer cells (30). Furthermore, a recent study has identified 3688 abnormal alternative splicing events in LUAD. Seven types of alternative splicing events were identified, with SKIP being most significant type in LUAD (30). In this study, we used the ASprofile software to categorize variable splicing types into 12 classes: TSS, TTS, SKIP, XSKIP, MSKIP, XMSKIP, IR, XIR, MIR, XMIR, AE, and XAE (Figure 7A). Then, we compared of alternative splicing between NM and LC, and found that the XMIR, XIR, XAE, MIR, and IR events were significantly upregulated in LUAD tissues compared to normal tissues (Figure 7B). These findings deepen our understanding of certain splicing events in LUAD and provide new targets and strategies for further study of the diagnosis and treatment of this disease.
Discussion
As a malignant tumor, LUAD’s occurrence and development are regulated by various complex molecular mechanisms. TFs play a crucial role in LUAD by regulating multiple key genes and signaling pathways, influencing the occurrence, development, and transformation of LUAD. However, the regulatory mechanisms of TFs in LUAD are highly complex. Despite previous studies identifying several important TFs in LUAD, many key TFs remain unexplored, and no research has been conducted on constructing TF-target gene regulatory networks based on the differentially expressed genes in LUAD. Therefore, in this study, we further identified key TFs in LUAD through high-throughput sequencing and, for the first time, constructed a key TF-target gene regulatory network based on the differentially expressed genes in LUAD. Interestingly, during the exploration of target genes regulated by key TFs, we discovered a potential oncogenic TF (GRHL3) that may regulate the Wnt signaling pathway, as well as a potential tumor-suppressive TF (TAL1) that may regulate signaling pathways associated with angiogenesis.
In this study, we first detected a large number of different expressed genes in human LUAD tissues through high-throughput sequencing, and these genes were significantly enriched in the Wnt and angiogenesis-related signaling pathways. In LUAD, the Wnt signaling pathway is frequently aberrantly activated, leading to the enhanced proliferation and invasive capabilities of LUAD cells, and is involved in the epithelial-mesenchymal transition (EMT) process and the maintenance and proliferation of LUAD stem cells (32,33). Angiogenesis also plays a critical role in various aspects of LUAD (34-36). First, angiogenesis provides the needed oxygen and nutrients to tumors, promoting the proliferation and growth of tumor cells. Second, the newly formed blood vessels provide the channels required for invasion and metastasis of LUAD cells. Moreover, angiogenesis contributes to therapeutic resistance in LUAD, as the newly formed blood vessels can reduce the effectiveness of drugs by providing alternative routes and lowering drug concentration (35). Therefore, the Wnt signaling pathway and angiogenesis are considered two important therapeutic targets in LUAD.
GRHL3 is a gene that encodes a protein belonging to the grainyhead family. It plays a crucial role in regulating biological processes such as epithelial cell differentiation, proliferation, and migration (37). Aberrant expression of GRHL3 in tumors is closely associated with the occurrence, development, and prognosis of various cancers. For example, GRHL3 can suppress squamous cell carcinoma by regulating the activation of PTEN/PI3K/Akt pathway (38). It exerts anticancer effects in esophageal squamous cell carcinoma by regulating HOPX expression and the Wnt signaling pathway (23). In addition, it promotes tumor growth and metastasis in colorectal cancer through the MEK pathway (39). Thus, related studies have largely shown the tumor-suppressive role of GRHL3, with a few cases demonstrating its oncogenic potential. This dual role of GRHL3 in tumors may be attributed to its transcriptional regulation of different target genes. For instance, GRHL3 can enhance the expression of the tumor suppressors PTEN or HOPX, thereby inhibiting the progression of esophageal squamous cell carcinoma (23,38). However, to date, no studies have revealed the specific regulatory target genes responsible for the oncogenic function of GRHL3, and there is limited research on the function of GRHL3 in lung cancer. In this study, we observed upregulated expression of GRHL3 in LUAD and found a negative correlation between GRHL3 expression and the survival rate of patients with lung cancer. Through target gene prediction and analysis using multiple databases, we identified multiple binding sites of GRHL3 in the promoter region of CDH15, suggesting CDH15 as a potential target gene of GRHL3. CDH15, also known as M-cadherin, belongs to the cadherin superfamily of calcium-dependent cell adhesion proteins and figures prominently in regulating the arrangement and fusion of myocytes (24). Existing research indicates that CDH15 may be associated with the occurrence, progression, and prognosis of certain tumors. For example, high methylation of CDH15 is correlated with poor survival in patients with liver cancer (40). Overall, CDH15 is considered to be a factor that promotes tumor cell invasion and metastasis. A study has suggested that the M-cadherin-mediated signaling pathway attenuates the phosphorylation of β-catenin (Ser31/37/Thr41) by GSK-3β (24). Therefore, we hypothesize that GRHL3 exerts its oncogenic function by transcriptionally regulating CDH15 expression and modulating the Wnt/β-catenin signaling pathway.
TAL1 is a gene that regulates hematopoietic development by controlling the transcription of multiple genes, including GATA1, GATA2, and LMO2 (41). Abnormalities in the TAL1 gene are common in T-cell acute lymphocytic leukemia (T-ALL), a malignant tumor occurring in children and adults (25). In patients with T-ALL, the TAL1 gene often undergoes chromosomal translocations, rearrangements, and mutations, leading to overexpression of the TAL1 protein (25). This disruption may interfere with normal hematopoietic differentiation processes and promote abnormal proliferation and survival of leukemia cells. However, there is limited research on the role of TAL1 in other malignancies, particularly in LUAD. In this study, we observed the significant downregulation of TAL1 expression in LUAD and found a positive correlation between TAL1 expression and the survival rate of patients with LUAD. Further analysis revealed that the potential target genes of TAL1 in LUAD were significantly enriched in the pathways related to vascular smooth muscle contraction and vascular regulation, which is consistent with the known functions of TAL1. Additionally, we identified ADAMTS1 as a candidate target gene of TAL1 in LUAD. ADAMTS1 showed a consistent expression pattern with TAL1 in LUAD tissues, and its promoter region contains multiple binding sites for TAL1. ADAMTS1 is involved not only in vascular development and angiogenesis but also in vascular growth and repair processes through its regulation of endothelial cell proliferation and migration (27). Furthermore, ADAMTS1 has been found to regulate vascular constriction and dilation by affecting the function of endothelial and smooth muscle cells, thus impacting vascular contractility and hemodynamics (27). These findings suggest that ADAMTS1 may be a target gene of TAL1 in LUAD. Therefore, we hypothesize that TAL1 may exert its anticancer effects by transcriptionally regulating the expression of ADAMTS1, thereby modulating vascular formation or contraction in LUAD.
Alternative splicing is an important gene expression regulatory mechanism that allows a single gene to produce multiple distinct transcripts and protein isoforms. Alternative splicing plays a crucial regulatory role in regulating the Wnt signaling pathway. β-catenin is a core regulatory factor in the Wnt signaling pathway, involved in gene transcription and cell fate determination. Alternative splicing can affect the expression levels and functions of β-catenin. For example, certain tumors exhibit alternatively spliced variants of β-catenin that may display enhanced transcriptional activity and tumor-promoting effects (42). Additionally, alternative splicing is critically involved in angiogenesis, affecting the expression and regulation of multiple genes involved in vascular formation and function (43). For instance, the precursor mRNA of key regulators in angiogenesis, such as members of the VEGF family including VEGF-A, VEGF-B, VEGF-C, and VEGF-D, undergoes alternative splicing to generate multiple transcript variants encoding proteins with distinct functions (44). These alternative splicing variants can regulate the stability, activity, and receptor-binding capacity of VEGF family members, thereby influencing endothelial cell proliferation, migration, and vascular formation (44). To further identify the transcriptional regulatory mechanisms in LUAD, we performed transcriptome sequencing and analyzed changes in alternative splicing events in LUAD. We found that XMIR, XIR, XAE, MIR, and IR events were significantly upregulated in LUAD tissues compared to normal tissues, suggesting that these five alternative splicing events exert meaningful regulatory roles in the Wnt and vascular-related pathways during the development and progression of LUAD.
In addition, abnormal regulation of cell proliferation, immune evasion, and EMT are also associated with the occurrence and development of LUAD as a malignant tumour (5,45). The major pathways enriched in TFs include not only transcriptional dysregulation in cancer, but also immune response-related pathways such as the TNF signaling pathway. The tumor immune microenvironment is the main source of heterogeneity in lung cancer, affecting disease progression and response to treatment. Heterogeneity and immune editing within genomic tumors are common early phenomena that may occur before invasion. Multi-region whole exome sequencing analysis revealed that mutations in EGFR, ERBB2, NRAS, and BRAF are early clonal genomic events in LUAD carcinogenesis, while TP53 and genes related to cell migration, gap junctions, and metastasis may be late events associated with sub-clonal diversity and tumor progression (46). A latest study utilized a highly reusable imaging quality control cytometer (IMC) to analyze the spatial resolution features related to lung tumour immune microenvironment and clinical outcomes in LUAD patients (47). The results showed that AI based systems can extract features from raw IMC images to predict various LUAD clinical outcomes. We will continue to investigate TFs and their regulatory networks associated with the immune microenvironment in LUAD patients.
Conclusions
In summary, we used transcriptomic sequencing to construct a regulatory network of key TFs and target genes involved in LUAD. Two potential key TFs involved in the regulation of LUAD (GRHL3 and TAL1) were identified, along with five alternative splicing events (XMIR, XIR, XAE, MIR, and IR) that exhibited abnormal occurrence in LUAD. We propose the existence of a GRHL3-CDH15-Wnt-β-catenin pro-oncogenic signaling axis and a TAL1-ADAMTS1-vascular antioncogenic signaling axis. Although further experimental validation is required to verify these complex transcriptional networks and pro- and antioncogenic signaling axes, this study provides potential targets and clues for the diagnosis and treatment of LUAD in the future.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1688/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1688/dss
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1688/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1688/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Ethics Committee of Binhai County People’s Hospital of Yancheng (No. 2021BHKYLL0023) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wu F, Wang L, Zhou C. Lung cancer in China: current and prospect. Curr Opin Oncol 2021;33:40-6. [Crossref] [PubMed]
- Jiang M, Qi F, Zhang K, et al. MARCKSL1-2 reverses docetaxel-resistance of lung adenocarcinoma cells by recruiting SUZ12 to suppress HDAC1 and elevate miR-200b. Mol Cancer 2022;21:150. [Crossref] [PubMed]
- Bade BC, Dela Cruz CS. Lung Cancer 2020: Epidemiology, Etiology, and Prevention. Clin Chest Med 2020;41:1-24. [Crossref] [PubMed]
- Liu L, Xu S, Huang L, et al. Systemic immune microenvironment and regulatory network analysis in patients with lung adenocarcinoma. Transl Cancer Res 2021;10:2859-72. [Crossref] [PubMed]
- Imyanitov EN, Iyevleva AG, Levchenko EV. Molecular testing and targeted therapy for non-small cell lung cancer: Current status and perspectives. Crit Rev Oncol Hematol 2021;157:103194. [Crossref] [PubMed]
- Hoy H, Lynch T, Beck M. Surgical Treatment of Lung Cancer. Crit Care Nurs Clin North Am 2019;31:303-13. [Crossref] [PubMed]
- Palazzo LL, Sheehan DF, Tramontano AC, et al. Disparities and Trends in Genetic Testing and Erlotinib Treatment among Metastatic Non-Small Cell Lung Cancer Patients. Cancer Epidemiol Biomarkers Prev 2019;28:926-34. [Crossref] [PubMed]
- Peng J, Yang S, Ng CSH, et al. The role of FOXP3 in non-small cell lung cancer and its therapeutic potentials. Pharmacol Ther 2023;241:108333. [Crossref] [PubMed]
- Sharma T, Das P, Panigrahi R, et al. Immunocytochemical Evaluation of TTF-1, Napsin-A, and p-63 for Subtyping of Non-Small Cell Lung Carcinoma and Clinicopathological Correlation. J Cytol 2022;39:180-7. [Crossref] [PubMed]
- Massó-Vallés D, Beaulieu ME, Soucek L. MYC, MYCL, and MYCN as therapeutic targets in lung cancer. Expert Opin Ther Targets 2020;24:101-14. [Crossref] [PubMed]
- Issaeva N, Bozko P, Enge M, et al. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med 2004;10:1321-8. [Crossref] [PubMed]
- West HJ, McCleland M, Cappuzzo F, et al. Clinical efficacy of atezolizumab plus bevacizumab and chemotherapy in KRAS-mutated non-small cell lung cancer with STK11, KEAP1, or TP53 comutations: subgroup results from the phase III IMpower150 trial. J Immunother Cancer 2022;10:e003027. [Crossref] [PubMed]
- Chen B, Gao S, Ji C, et al. Integrated analysis reveals candidate genes and transcription factors in lung adenocarcinoma. Mol Med Rep 2017;16:8371-9. [Crossref] [PubMed]
- Otálora-Otálora BA, López-Kleine L, Rojas A. Lung Cancer Gene Regulatory Network of Transcription Factors Related to the Hallmarks of Cancer. Curr Issues Mol Biol 2023;45:434-64. [Crossref] [PubMed]
- Yao Y, Luo J, Sun Q, et al. HOXC13 promotes proliferation of lung adenocarcinoma via modulation of CCND1 and CCNE1. Am J Cancer Res 2017;7:1820-34.
- Tang XL, Ding BX, Hua Y, et al. HOXC10 Promotes the Metastasis of Human Lung Adenocarcinoma and Indicates Poor Survival Outcome. Front Physiol 2017;8:557. [Crossref] [PubMed]
- Mimae T, Okada M, Hagiyama M, et al. Upregulation of notch2 and six1 is associated with progression of early-stage lung adenocarcinoma and a more aggressive phenotype at advanced stages. Clin Cancer Res 2012;18:945-55. [Crossref] [PubMed]
- Hou H, Yu X. Six2 promotes non-small cell lung cancer cell stemness via transcriptionally and epigenetically regulating E-cadherin. Cell Prolif 2019;52:e12617. [Crossref] [PubMed]
- Wang Y, Ding X, Liu B, et al. ETV4 overexpression promotes progression of non-small cell lung cancer by upregulating PXN and MMP1 transcriptionally. Mol Carcinog 2020;59:73-86. [Crossref] [PubMed]
- Wang Z, Wei Y, Zhang R, et al. Multi-Omics Analysis Reveals a HIF Network and Hub Gene EPAS1 Associated with Lung Adenocarcinoma. EBioMedicine 2018;32:93-101. [Crossref] [PubMed]
- Hedrick E, Mohankumar K, Safe S. TGFβ-Induced Lung Cancer Cell Migration Is NR4A1-Dependent. Mol Cancer Res 2018;16:1991-2002. [Crossref] [PubMed]
- Zhang Q, Liu W, Zhang HM, et al. hTFtarget: A Comprehensive Database for Regulations of Human Transcription Factors and Their Targets. Genomics Proteomics Bioinformatics 2020;18:120-8. [Crossref] [PubMed]
- Georgy SR, Rudiatmoko DR, Auden A, et al. Identification of a Novel GRHL3/HOPX/Wnt/β-Catenin Proto-oncogenic Axis in Squamous Cell Carcinoma of the Esophagus. Cell Mol Gastroenterol Hepatol 2023;15:1051-69. [Crossref] [PubMed]
- Wang Y, Mohamed JS, Alway SE. M-cadherin-inhibited phosphorylation of ß-catenin augments differentiation of mouse myoblasts. Cell Tissue Res 2013;351:183-200. [Crossref] [PubMed]
- Porcher C, Chagraoui H, Kristiansen MS. SCL/TAL1: a multifaceted regulator from blood development to disease. Blood 2017;129:2051-60. [Crossref] [PubMed]
- Deleuze V, Chalhoub E, El-Hajj R, et al. TAL-1/SCL and its partners E47 and LMO2 up-regulate VE-cadherin expression in endothelial cells. Mol Cell Biol 2007;27:2687-97. [Crossref] [PubMed]
- Iruela-Arispe ML, Carpizo D, Luque A. ADAMTS1: a matrix metalloprotease with angioinhibitory properties. Ann N Y Acad Sci 2003;995:183-90. [Crossref] [PubMed]
- Tan Ide A, Ricciardelli C, Russell DL. The metalloproteinase ADAMTS1: a comprehensive review of its role in tumorigenic and metastatic pathways. Int J Cancer 2013;133:2263-76. [Crossref] [PubMed]
- Yin D, Lin D, Xie Y, et al. Neuregulin-1β Alleviates Sepsis-Induced Skeletal Muscle Atrophy by Inhibiting Autophagy via AKT/mTOR Signaling Pathway in Rats. Shock 2022;57:397-407. [Crossref] [PubMed]
- Coomer AO, Black F, Greystoke A, et al. Alternative splicing in lung cancer. Biochim Biophys Acta Gene Regul Mech 2019;1862:194388. [Crossref] [PubMed]
- Pio R, Montuenga LM. Alternative splicing in lung cancer. J Thorac Oncol 2009;4:674-8. [Crossref] [PubMed]
- Stewart DJ. Wnt signaling pathway in non-small cell lung cancer. J Natl Cancer Inst 2014;106:djt356. [Crossref] [PubMed]
- Krishnamurthy N, Kurzrock R. Targeting the Wnt/beta-catenin pathway in cancer: Update on effectors and inhibitors. Cancer Treat Rev 2018;62:50-60. [Crossref] [PubMed]
- Sridhar SS, Shepherd FA. Targeting angiogenesis: a review of angiogenesis inhibitors in the treatment of lung cancer. Lung Cancer 2003;42:S81-91. [Crossref] [PubMed]
- Shukla NA, Yan MN, Hanna N. The Story of Angiogenesis Inhibitors in Non-small-cell Lung Cancer: The Past, Present, and Future. Clin Lung Cancer 2020;21:308-13. [Crossref] [PubMed]
- Eelen G, Treps L, Li X, et al. Basic and Therapeutic Aspects of Angiogenesis Updated. Circ Res 2020;127:310-29. [Crossref] [PubMed]
- Bai Y, Zhao Z, Boath J, et al. The functional GRHL3-filaggrin axis maintains a tumor differentiation potential and influences drug sensitivity. Mol Ther 2021;29:2571-82. [Crossref] [PubMed]
- Darido C, Georgy SR, Wilanowski T, et al. Targeting of the tumor suppressor GRHL3 by a miR-21-dependent proto-oncogenic network results in PTEN loss and tumorigenesis. Cancer Cell 2011;20:635-48. [Crossref] [PubMed]
- Tan L, Qu W, Wu D, et al. GRHL3 Promotes Tumor Growth and Metastasis via the MEK Pathway in Colorectal Cancer. Anal Cell Pathol (Amst) 2021;2021:6004821. [Crossref] [PubMed]
- Yamada S, Nomoto S, Fujii T, et al. Frequent promoter methylation of M-cadherin in hepatocellular carcinoma is associated with poor prognosis. Anticancer Res 2007;27:2269-74.
- Vagapova ER, Spirin PV, Lebedev TD, et al. The Role of TAL1 in Hematopoiesis and Leukemogenesis. Acta Naturae 2018;10:15-23.
- Sumithra B, Saxena U, Das AB. Alternative splicing within the Wnt signaling pathway: role in cancer development. Cell Oncol (Dordr) 2016;39:1-13. [Crossref] [PubMed]
- Bowler E, Oltean S. Alternative Splicing in Angiogenesis. Int J Mol Sci 2019;20:2067. [Crossref] [PubMed]
- Biselli-Chicote PM, Oliveira AR, Pavarino EC, et al. VEGF gene alternative splicing: pro- and anti-angiogenic isoforms in cancer. J Cancer Res Clin Oncol 2012;138:363-70. [Crossref] [PubMed]
- Zhu C, Xie Y, Li Q, et al. CPSF6-mediated XBP1 3’UTR shortening attenuates cisplatin-induced ER stress and elevates chemo-resistance in lung adenocarcinoma. Drug Resist Updat 2023;68:100933. [Crossref] [PubMed]
- Zhang C, Zhang J, Xu FP, et al. Genomic Landscape and Immune Microenvironment Features of Preinvasive and Early Invasive Lung Adenocarcinoma. J Thorac Oncol 2019;14:1912-23. [Crossref] [PubMed]
- Sorin M, Rezanejad M, Karimi E, et al. Single-cell spatial landscapes of the lung tumour immune microenvironment. Nature 2023;614:548-54. [Crossref] [PubMed]


