Mutation profile in liquid biopsy tested by next generation sequencing in Mexican patients with non-small cell lung carcinoma and its impact on survival
Highlight box
Key findings
• 32.3% of patients with negative epidermal growth factor receptor via reverse transcription-polymerase chain reaction maintain a target mutation to guide treatment.
• Next-generation sequencing (NGS) has a positive impact on progression-free survival (PFS) and overall survival (OS).
What is known and what is new?
• NGS allows physicians to identify specific mutations in a certain tumor and use targeted drugs, as studied in multiple trials such as the NCI-MPACT, MOSCATO-01, and MOST trials.
• Our study is the first descriptive and analytical study carried out in Mexican population correlating specific molecular mutations in patients with non-small cell lung cancer using NGS from liquid biopsy sample via a 62-gene panel and correlating their impact on PFS and OS.
What is the implication, and what should change now?
• NGS must be implemented in developing countries to guide treatment.
Introduction
Background
Lung cancer, the leading cause of cancer-related deaths worldwide, accounted for an estimated 1,796,144 fatalities in 2020 (1). In the United States, there were an estimated 227,875 new cases in 2020, with 138,225 estimated deaths, resulting in a 5-year relative survival rate of 21.7%. In contrast, Mexico reported 7,588 new cases, along with 7,100 deaths, and a 5-year survival rate of 6.37% (1,2). Regrettably, lung cancer is frequently diagnosed at an advanced stage, leading to a bleak prognosis, and treatment typically relies on multiple chemotherapeutic regimens. Recent advancements in gene sequencing, facilitated by next-generation sequencing (NGS) and immunotherapy, have paved the way for personalized treatments, with the ultimate aim of enhancing overall survival (OS).
The inaugural nucleic acid sequencing, known as the Sanger method, was developed in 1970. Nowadays, cutting-edge technologies are referred to as NGS, which enables billions of sequencing reactions to occur simultaneously (3-5). The results are mapped to online genome databases, and specific genetic mutations are sought after and compared. NGS can sequence the entire genome within 24 hours, thus revolutionizing the field of oncology (6). NGS empowers physicians to pinpoint specific mutations in a given tumor and employ targeted drugs, as demonstrated in various trials such as NCI-MPACT, MOSCATO-01, and MOST (7).
While NGS is evolving into a valuable tool, it does come with certain limitations, including high costs, data interpretation challenges, and a limited amount of tumor sample obtained from the initial biopsy, often necessitating rebiopsy, which is not always feasible (7). In such instances, liquid biopsy emerges as the sole option to detect molecular targets, particularly in non-small cell lung cancer (NSCLC) patients. It’s crucial to note that the gold standard for lung cancer diagnosis remains tumor biopsy. Therefore, combining liquid biopsy with NGS has secured Food and Drug Administration (FDA) approval as a companion diagnostic test for patients with NSCLC, assisting in guiding treatment decisions (8-10). The analyte employed by NGS for liquid biopsies is circulating tumor DNA (ctDNA), extracted from plasma, and is the sole clinically approved analyte. It boasts high specificity and has undergone extensive study, particularly for assessing epidermal growth factor receptor (EGFR) status (11,12).
Rationale and knowledge gap
The combined use of NGS and liquid biopsy has primarily found application in the study of advanced-stage lung cancer, as plasma levels of ctDNA are more pronounced in these stages. Nevertheless, it has also been explored for screening and early detection in individuals at high risk to ensure appropriate treatment (11,13,14). In the context of EGFR mutations, liquid biopsy stands as a valid alternative to tissue samples for molecular testing, with the goal of administering tyrosine kinase inhibitors (TKIs). In fact, its application has yielded an overall response rate of 72% and a median progression-free survival (PFS) of 11 months (15,16). This approach also proves beneficial in cases where patients undergoing TKI treatment develop resistance mechanisms, especially through the emergence of the T790M mutation (17). The ongoing BFAST multi-cohort study, which assesses the relationship between NGS-based detection of anaplastic lymphoma kinase (ALK) mutations in blood and the effectiveness of targeted therapies, has substantiated the clinical advantages of ALK fusions in patients receiving alectinib, thus establishing NGS as a valuable tool for therapeutic decision-making in patients with advanced NSCLC and ALK mutations (18).
The therapeutic decision-making process relies on the identification of biomarkers in either tissue or blood samples. In the pursuit of streamlining and optimizing treatment, the European Society for Medical Oncology Scale of Clinical Actionability of Molecular Targets (ESCAT) was developed, classifying DNA mutations based on evidence-based medicine. Given the recent discovery of new targeted therapies tailored to specific genetic mutations in NSCLC management, the ESCAT classification system, initially published in 2018, calls for an update to incorporate the insights offered by this classification. This article serves as an update on the ESCAT level 1 driver mutations identified within our population.
Objectives
- To delineate the mutational profile of patients diagnosed with NSCLC in the Mexican population through NGS.
- To perform an analysis of PFS and OS in patients diagnosed with advanced mutated adenocarcinoma NSCLC.
We present this article in accordance with the REMARK reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1029/rc).
Methods
This was a descriptive, observational, retrospective study aimed at delineating the molecular mutations observed in patients with NSCLC of the adenocarcinoma subtype through NGS analysis of liquid biopsy samples obtained from Mexican patients, our group was able to access NGS via liquid biopsy through a financial requirement access, which did not include tissue evaluation by NGS. The study was conducted at the Instituto Nacional de Enfermedades Respiratorias – Ismael Cosio Villegas, located in Mexico City, within the Department of Thoracic Oncology. A comprehensive review of electronic clinical records spanning the last 5 years resulted in the identification of a total of 127 patients. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Institutional Ethics Board Committee “Comité de Ética en Investigación Instituto Nacional de Enfermedades Respiratorias” (E08-19) and informed consent was taken from all the patients.
Patients with a confirmed NSCLC diagnosis were assessed through liquid biopsy samples, offering them the opportunity to receive targeted therapy based on the results. Before conducting a liquid biopsy, we determined the EGFR mutation status by analyzing tumor tissue samples with the Idylla™ real-time PCR method, using the Idylla™ EGFR Mutation Test. Patients without detectable EGFR mutations were subsequently subjected to liquid biopsy analysis via NGS prior to the initiation of first-line therapy. Our study concentrated on the molecular insights derived from the liquid biopsy results, which were processed using NGS technology provided by Foundation Medicine, a pharmaceutical and molecular diagnostics company. This comprehensive analysis explored more than 60 genes (Box 1), detecting a range of mutations, including insertions, deletions, and base substitutions.
| ABL1 | AKT1 | ALK | ARAF | BRAF |
| BRCA1 | BRCA2 | BTK | CCND1 | CD274 |
| CDH1 | CDK4 | CDK6 | CDKN2A | CRKL |
| CTNNB1 | DDR2 | EGFR | ERBB2 | ERRFI1 |
| ESR1 | EZH2 | FGFR1 | FGFR2 | FGFR3 |
| FLT3 | FOXL2 | GNA11 | GNAQ | GNAS |
| HRAS | IDH1 | IDH2 | JAK2 | JAK3 |
| KIT | KRAS | MAP2K1 | MAP2K2 | MDM2 |
| MET | MPL | MTOR | MYC | MYCN |
| MYD88 | NF1 | NPM1 | NRAS | PDCD1LG2 |
| PDGFRA | PDGFRB | PIK3CA | PTEN | PTPN11 |
| RAF1 | RET | ROS1 | SMO | TERT |
| TP53 | VEGFA |
Statistical analysis
Statistical analysis encompassed a descriptive examination of categorical variables, with their values expressed in terms of number and percentage. These variables included gender, age, smoking status, exposure to wood smoke, asbestos exposure, chronic obstructive pulmonary disease (COPD), type 2 diabetes mellitus, systemic arterial hypertension, Eastern Cooperative Oncology Group (ECOG) performance status, stage of diagnosis, presence of brain and liver metastases, clinical presentation, and mutation type. The impact of mutation type on PFS and OS was determined by calculating 95% confidence intervals (CIs) and P values. A multivariate logistic regression analysis was conducted. Interferential comparisons were facilitated through Student’s t-distribution or Mann-Whitney U tests, depending on the data distribution as determined by the Kolmogorov-Smirnov test. The chi-square and Fisher’s exact tests were utilized to assess the statistical significance of each categorical variable.
Survival analyses were performed using the log-rank test to compare outcomes between groups, with results deemed statistically significant if P≤0.05. Variables found to be significant or of borderline significance were integrated into the logistic regression analysis if the relationship exceeded a 10% threshold. Treatment response was evaluated according to Response Evaluation Criteria in Solid Tumours version 1.1 (RECIST v1.1) criteria and expressed as a percentage. PFS was calculated as the time elapsed from the initiation of treatment to the date of progression, while OS was measured from the date of diagnosis to the date of death. These survival analyses were conducted using the Kaplan-Meier technique, and group comparisons were made through the log-rank test. The relative risk for each profile was calculated, with the profile exhibiting the highest risk ratio being selected. Data analysis was performed using SPSS version 19 software.
Results
Population characteristics
A total of 127 patients diagnosed with advanced NSCLC, specifically of the adenocarcinoma subtype, were included in the analysis. These patients were treated at a Mexican national reference cancer center, and they all tested negative for the classic EGFR mutation via reverse transcription-polymerase chain reaction (rtPCR). Subsequently, they underwent NGS analysis using liquid biopsy samples. Among the entire population, there were 60 male patients (47.2%) and 67 females (52.8%), with an average age of 60 years, ranging from 27 to 87 years. The majority of patients were smokers (52.8%), with an average smoking index of 20.57±28.08. Additionally, 32.3% of patients had been exposed to asbestos, and 46.5% had a history of wood smoke exposure. In terms of associated comorbidities, only 9.9% had been diagnosed with COPD, 11.8% had type 2 diabetes mellitus, and 29.1% had chronic hypertension.
At the time of diagnosis, the functional status of the patients, as measured by the ECOG scale, indicated that 89.0% had an ECOG status of 0–1, while 11% had an ECOG status of 2. At the time of diagnosis, 100 patients were classified as stage IV (91.3%), and 17 patients (13.4%) presented with brain metastases, while 8 patients (6.3%) had liver metastases. Lung cancer patients exhibited a wide range of clinical symptoms; in this population, 78.7% presented with cough, followed by dyspnea (59.1%), chest pain (40.2%), weight loss (23.6%), and hemoptysis (15.7%). The complete demographic data for the population is presented in Table 1.
Table 1
| Characteristics | Values |
|---|---|
| Gender, n (%) | |
| Male | 60 (47.2) |
| Female | 67 (52.8) |
| Age, years, mean ± SD [range] | 60.9±12.7 [27–87] |
| Smoking status | |
| Never-smoker, n (%) | 60 (47.2) |
| Smoker, n (%) | 67 (52.8) |
| Smoking index (pack years), mean ± SD | 20.57±28.08 |
| Exposure to wood smoke, n (%) | 59 (46.5) |
| Asbestos exposure, n (%) | 41 (32.3) |
| Comorbidities, n (%) | |
| COPD | 12 (9.9) |
| T2D | 15 (11.8) |
| SAH | 37 (29.1) |
| ECOG, n (%) | |
| 0–1 | 113 (89.0) |
| 2 | 14 (11.0) |
| Stage, n (%) | |
| II–III | 11 (8.7) |
| IV | 116 (91.3) |
| Brain metastases, n (%) | 17 (13.4) |
| Liver metastases, n (%) | 8 (6.3) |
| Clinical characteristic, n (%) | |
| Chest pain | 51 (40.2) |
| Dyspnea | 75 (59.1) |
| Weight loss | 30 (23.6) |
| Cough | 100 (78.7) |
| Hemoptysis | 20 (15.7) |
SD, standard deviation; COPD, chronic obstructive pulmonary disease; T2D, type 2 diabetes; SAH, systemic arterial hypertension; ECOG, Eastern Cooperative Oncology Group.
Genetic mutations identified
One of the primary objectives of this study was to identify the most common mutations in patients, which were analyzed via NGS before the initiation of targeted therapy. Out of the 127 patients, 22 (17.3%) did not have an identifiable mutation. Among these 22 cases, 7 patients had variants of uncertain significance (VUS), while 15 patients had no identifiable mutations at all. The most prevalent mutation identified was EGFR, found in 24 patients (18.9%), followed by Kirsten rat sarcoma viral oncogene homolog (KRAS) in 20 patients (15.7%), tumor protein 53 (TP53) as the primary target mutation in 13 patients (10.2%), ALK in 11 patients (8.7%), and p110α subunit of phosphatidylinositol 3-kinases (PIK3CA) in 6 patients (4.7%). Other significant mutations included rearranged during transfection (RET) in 5 patients (3.9%), v-raf murine sarcoma viral oncogene homolog B1 (BRAF) in 4 patients (3.1%), breast cancer (BRCA)1/2 gene and human epidermal growth factor receptor 2 (HER2) in 3 patients each (2.4%), and cyclin dependent kinase inhibitor 2A (CDKN2A), isocitrate dehydrogenase 2 (IDH2), mesenchymal epithelial transition factor (MET), and myelocytomatosis (MYC), each identified in 2 patients (1.6%). Additional mutations, such as ‘orphan’ c-ros oncogene 1 (ROS1), telomerase reverse transcriptase (TERT), neuroblastoma RAS viral oncogene homolog (NRAS), neurofibromin type 1 (NF1), Janus kinase 2 (JAK2) gene, fibroblast growth factor receptor 2 (FGFR2), FGFR3, and fms-like tyrosine kinase 3 (FLT3), were each identified in a single patient (0.8%). A summary of all the mutations identified in the population can be found in Figure 1.
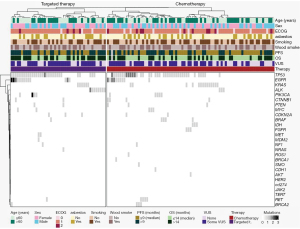
Thirteen patients were identified with TP53 as the primary target mutation, while a total of 65 patients exhibited TP53 mutations as co-mutations. From our population, 46 patients (36.2%) received targeted therapy, 69 patients (54.33%) received chemotherapy, and 12 cases (9.45%) did not receive any treatment. Patients who received targeted therapy did so from the moment of diagnosis and throughout the course of their disease (Figure 2).
Factors influencing PFS
The overall median PFS was 9 months (95% CI, 8.34–9.65) (Figure 3). Patients without any identified mutations had a median PFS of 8 months (95% CI, 7.08–8.91). Median PFS in months was analyzed for each identified mutation. ALK mutations exhibited the longest PFS at 23 months (95% CI, 18.68–27.31), followed by FGFR with 19 months (NA), EGFR with 15 months (95% CI, 13.12–16.87), and MYC with 11 months (NA). BRCA (95% CI, 5.19–14.80), BRAF (95% CI, 5.75–14.24), and RET (95% CI, 3.61–12.38) displayed a median PFS of 10 months, with KRAS (95% CI, 6.95–9.04), TERT (NA), JAK2 (NA), IDH2 (NA), and HER2 (NA) following with an 8-month PFS. The mutations with lower PFS included TP53 (95% CI, 1.34–10.52) and PIK3CA (95% CI, 1.34–12.65) with a 7-month PFS, while MET (NA) and FLT3 (NA) had a 6-month PFS, and NF1 (NA) a 5-month PFS, with NRAS (NA) displaying a 4-month PFS.
Notably, statistically significant differences were observed among the median PFS of mutations, with ALK, EGFR, KRAS, FGFR, and MYC being the most statistically significant. Worth mentioning, eight out of the eleven patients with ALK mutations received targeted therapy, and 14 out of 24 patients with EGFR mutations received the same treatment.
From the variables under study, a univariate analysis was conducted to identify the statistically significant variables for PFS. Subsequently, a multivariate analysis was performed, which included COPD, ECOG, brain MRI, tumor mutation burden (TMB), ALK, EGFR, KRAS, FGFR, MYC, and protein kinase B (AKT) (as an identified co-mutation, not a target mutation).
Although the previously mentioned variables yielded significant P values in the univariate analysis, ALK [hazard ratio (HR): 0.258, P<0.001], EGFR (HR: 0.538, P=0.041), TMB (HR: 2.073, P=0.042) and brain MRI (HR: 0.470, P=0.032) retained statistical significance in the multivariate analysis. Additionally, ESCAT Level 1 mutations identified in our population included KRAS (HR: 1.37, P=0.24), while MET, RET, and BRAF did not reach significance in the univariate statistical analysis, potentially due to the relatively small population sample. The complete list of the univariate and multivariate analyses for each factor concerning PFS is provided in Table 2.
Table 2
| Characteristics/factors | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| Median (95% CI) | P value† | Hazard ratio | P value‡ | ||
| Overall | 9 (8.34–9.65) | ||||
| No. of pathogenic mutations | – | 0.21 | 1.164 (0.970–1.397) | 0.10 | |
| Target therapy | – | 0.21 | 0.798 (0.559–1.141) | 0.21 | |
| COPD | – | 0.04 | 1.050 (0.483–2.283) | 0.90 | |
| ECOG | – | 0.04 | 1.315 (0.851–2.033) | 0.21 | |
| Brain MRI | – | 0.03 | 0.470 (0.236–0.938) | 0.032 | |
| TMB | – | 0.01 | 2.073 (1.025–4.191) | 0.042 | |
| AKT | – | 0.01 | 0.671(0.068–6.602) | 0.73 | |
| KRAS | 8 (6.95–9.04) | 0.04 | 1.37 (0.810–2.32) | 0.24 | |
| ALK | 23 (18.68–27.31) | <0.001 | 0.258 (0.126–0.53) | <0.001 | |
| BRCA | 10 (5.19–14.80) | 0.19 | – | – | |
| ROS1 | 10 (NA) | 0.42 | – | – | |
| EGFR | 15 (13.12–16.87) | 0.06 | 0.538 (0.297–0.975) | 0.041 | |
| MYC | 11 (NA) | 0.01 | 0.463 (0.194–1.1) | 0.081 | |
| NRAS | 4 (NA) | 0.20 | – | – | |
| BRAF | 10 (5.75–14.24) | 0.47 | – | – | |
| TP53 | 7 (1.34–10.52) | 0.40 | – | – | |
| RET | 10 (3.61–12.38) | 0.50 | – | – | |
| PIK3CA | 7 (1.34–12.65) | 0.49 | – | – | |
| NF1 | 5 (NA) | 0.19 | – | – | |
| FGFR | 19 (NA) | 0.005 | 0.878 (0.323–2.39) | 0.79 | |
| MET | 6 (NA) | 0.10 | – | – | |
| TERT | 8 (NA) | 0.34 | – | – | |
| JAK2 | 8 (NA) | 0.34 | – | – | |
| IDH2 | 8 (NA) | – | – | – | |
| HER2 | 8 (NA) | 0.28 | – | – | |
| FLT3 | 6 (NA) | – | – | – | |
†, Pearson’s correlation; ‡, Cox regression. NA, not available due to sample including only one patient. PFS, progression-free survival; CI, confidence interval; COPD, chronic obstructive pulmonary disease; ECOG, Eastern Cooperative Oncology Group; MRI, magnetic resonance imaging; TMB, Tumor Mutational Burden; AKT, protein kinase B; KRAS, Kirsten rat sarcoma viral oncogene homolog; ALK, anaplastic lymphoma kinase; BRCA, BReast CAncer gene; ROS1, ‘orphan’ receptor tyrosine kinase; EGFR, epidermal growth factor receptor; MYC, myelocytomatosis; NRAS, neuroblastoma RAS viral oncogene homolog; BRAF, v-raf murine sarcoma viral oncogene homolog B1; TP53, tumor protein 53; RET, rearranged during transfection; PIK3CA, p110α subunit of Phosphatidylinositol 3-kinases; NF1, Neurofibromin type 1; FGFR, fibroblast growth factor receptor; MET, mesenchymal epithelial transition factor; TERT, Telomerase reverse transcriptase; JAK2, Janus Kinase 2 gene; IDH2, isocitrate dehydrogenase 2; HER2, human epidermal growth factor receptor 2; FLT3, fms-like tyrosine kinase 3.
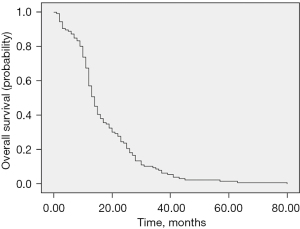
Factors influencing OS
The OS for the studied population stood at 14 months (95% CI, 12.43–15.56), boasting an impressive 1-year survival rate of 70% (Figure 4). Patients without identifiable mutations exhibited a median OS of 12 months (95% CI, 10.85–13.14). Among the analyzed mutations, ALK mutation took the lead with the highest median OS, standing at 35 months (95% CI, 25.28–44.71), closely followed by EGFR with 23 months (95% CI, 21.44–24.55). FGFR (NA) showed a respectable 19-month OS, while MYC (NA) demonstrated a commendable 16-month OS. Other mutations, such as BRCA (95% CI, 9.19–18.80), achieved a 14-month OS, whereas BRAF (95% CI, 6.14–19.86), IDH2 (NA), and HER2 (95% CI, 6.59–19.40) achieved a 13-month OS. NF1, JAK2, and KRAS displayed a 12-month OS, while TP53 and TERT were associated with an 11-month OS. On the lower end of the OS spectrum, FLT3 presented a 10-month OS, while PIK3CA exhibited a 9-month OS. MET, on the other hand, displayed a 7-month OS, and NRAS was associated with a 4-month OS. CDKN2A had a mere 2-month OS.
The same analytical approach, as employed in the analysis of PFS, was applied to OS. This comprehensive analysis included variables such as COPD, dyspnea, thoracic pain, ECOG, PET-CT, brain MRI, TMB, KRAS, ALK, EGFR, AKT, CTNNB1, and CD274 (with AKT, CTNNB1, and CD274 being identified as co-mutations rather than target mutations).
While all the aforementioned factors demonstrated statistical significance in the univariate analysis, ALK (HR: 0.285, P<0.001) and EGFR (HR: 0.482, P=0.024) retained statistical significance in the multivariate analysis. It’s essential to highlight those additional mutations found in our population, which hold significance, include KRAS (HR: 1.256, P=0.40). However, mutations like RET, MET, and BRAF did not achieve significance in the univariate analysis. It’s crucial to note that this lack of statistical significance may be attributed to the relatively small sample size of the population. For a complete list of factors with their respective univariate and multivariate analysis outcomes, please refer to Table 3.
Table 3
| Characteristics/factors | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| Median (95% CI) | P value† | Hazard ratio (95% CI) | P value‡ | ||
| Overall | 14 (12.43–15.56) | ||||
| Number of pathogenic mutations | – | 0.35 | 1.134 (0.916–1.404) | 0.24 | |
| Target therapy | – | 0.28 | 0.652 (0.446–0.952) | 0.027 | |
| COPD | – | 0.02 | 1.466 (0.654–3.29) | 0.35 | |
| Dyspnea | – | 0.02 | 1.373 (0.886–2.178) | 0.17 | |
| Thoracic Pain | – | 0.05 | 1.268 (0.840–1.912) | 0.25 | |
| ECOG | – | 0.01 | 1.398 (0.875–2.234) | 0.16 | |
| PET-CT | – | 0.07 | 0.859 (0.527–1.402) | 0.54 | |
| Brain MRI | – | 0.001 | 0.595 (0.272–1.303 | 0.19 | |
| TMB | – | 0.02 | 1.727 (0.834–3.575) | 0.14 | |
| AKT | – | 0.06 | 0.560 (0.057–5.507) | 0.61 | |
| CTNNB1 | – | 0.06 | 1.651 (0.539–5.057) | 0.38 | |
| CD274 | – | <0.001 | 0.491 (0.049–4.904) | 0.54 | |
| KRAS | 12 (7.61–16.38) | 0.04 | 1.256 (0.734–2.147) | 0.40 | |
| ALK | 35 (25.28–44.71) | <0.001 | 0.285 (0.141–0.574) | <0.001 | |
| BRCA | 14 (9.19–18.80) | 0.16 | – | – | |
| ROS1 | 11 | 0.30 | |||
| EGFR | 23 (21.44–24.55) | 0.009 | 0.482 (0.256–0.908) | 0.024 | |
| MYC | 16 (NA) | 0.12 | – | ||
| NRAS | 4 (NA) | 0.14 | – | – | |
| BRAF | 13 (6.14–19.86) | 0.30 | – | – | |
| TP53 | 11 (8.65–13.34) | 0.36 | – | – | |
| CDKN2A | 2 (NA) | 0.28 | – | – | |
| RET | 11 (0–25.03) | 0.30 | – | – | |
| PIK3CA | 9 (2.99–15.00) | 0.36 | – | – | |
| NF1 | 12 (NA) | 0.24 | – | – | |
| FGFR | 19 (NA) | 0.07 | 0.751 (0.289–1.952) | 0.55 | |
| MET | 7 (NA) | 0.26 | – | – | |
| TERT | 11 (NA) | 0.30 | – | – | |
| JAK2 | 12 (NA) | 0.33 | – | – | |
| IDH2 | 13 (NA) | – | – | – | |
| HER2 | 13 (6.59–19.40) | 0.45 | – | – | |
| FLT3 | 10 (NA) | – | – | – | |
†, Pearson’s correlation; ‡, Cox regression. NA, not available due to sample including only one patient. OS, overall survival; CI, confidence interval; COPD, chronic obstructive pulmonary disease; ECOG, Eastern Cooperative Oncology Group; MRI, magnetic resonance imaging; PET-CT, positron emission tomography/computed tomography; TMB, tumor mutational burden; AKT, protein kinase B; CTNNB1, catenin beta 1; CD274, PDL1; KRAS, Kirsten rat sarcoma viral oncogene homolog; ALK, anaplastic lymphoma kinase; BRCA, BReast CAncer gene; ROS1, ‘orphan’ receptor tyrosine kinase; EGFR, epidermal growth factor receptor; MYC, myelocytomatosis; NRAS, neuroblastoma RAS viral oncogene homolog; BRAF, v-raf murine sarcoma viral oncogene homolog B1; TP53, tumor protein 53; CDKN2A, cyclin dependent kinase inhibitor 2A, RET, rearranged during transfection; PIK3CA, p110α subunit of Phosphatidylinositol 3-kinases; NF1, Neurofibromin type 1; FGFR, fibroblast growth factor receptor; MET, mesenchymal epithelial transition factor; TERT, Telomerase reverse transcriptase; JAK2, Janus Kinase 2 gene; IDH2, isocitrate dehydrogenase 2; HER2, human epidermal growth factor receptor 2; FLT3, fms-like tyrosine kinase 3.
Updating the ESCAT classification system
A distinctive contribution of this paper is the revision of the ESCAT classification system for NSCLC. The ESCAT system was initially published in 2018 in the Annals of Oncology. Our revision stems from the identification of ESCAT level 1 driver mutations within our study population. In recent years, substantial advancements have been made in the management of NSCLC patients, particularly in the realm of innovative treatments designed for specific target mutations. As such, we view it as paramount to recognize mutations categorized as ESCAT level 1 based on the ESCAT definitions. To accomplish this, we drew upon the National Comprehensive Cancer Network (NCCN) guidelines, insights gleaned from recent phase 1, 2, and 3 clinical trials, and findings from high-impact publications. Furthermore, we collaborated with certified medical thoracic oncologists from the National Institute of Respiratory Diseases in Mexico City. Our aim is to provide fellow clinicians with up-to-date information about mutations that already have available targeted therapies to guide treatment decisions. We also emphasize that NGS can unveil previously unidentified target mutations when compared to traditional rtPCR methods.
It’s imperative to underline that ESCAT level 1 mutations are associated with validated drugs, providing a clear direction for treatment decisions. Our study population comprised 32.3% of patients harboring ESCAT level 1 mutations, despite previously testing negative for EGFR mutations through rtPCR. The mutations we identified encompassed a range of pathogenic mutations, including KRAS G12C, ALK fusion, RET fusion, EGFR deletion on exon 19, EGFR L858R, EGFR insertion on exon 20, MET splicing on exon 14, BRAF V600E, and ROS1 fusion. Each of these mutations corresponds to specific pathogenic alterations, which can be deletions, insertions, or fusions, that categorize them as ESCAT level 1 (IA or IB).
In our population, we identified 3 cases with the BRAF V600E mutation, this mutation is notable as it shows excellent responses to target therapy. A similar scenario unfolded in cases involving EGFR mutations. Out of the 24 EGFR-positive cases identified, only 14 received targeted therapy. This situation is lamentable, not just within our country, but in numerous developing countries, where many patients are unable to bear the financial burden of these drugs, which could otherwise offer a more promising prognosis.
While 36% of our patients received targeted therapy, and 32% were identified with ESCAT level 1 driver mutations, the higher percentage of cases receiving targeted treatment can be attributed to scenarios in which patients classified as ESCAT level 2 became participants in research protocols that granted them access to targeted therapies. Conversely, those with ESCAT level 1 driver mutations who did not receive targeted therapy were impeded by their inability to afford the treatment, along with limitations stemming from our institution.
Discussion
Lung cancer remains the fourth leading cause of cancer-related mortality in Mexico, and regrettably, it is diagnosed at an advanced stage in 68.5% of cases (1,19). The integration of biomarkers into lung cancer management for treatment, prognosis, and diagnosis has become indispensable. However, even in developed nations, not all recommended biomarkers, which include EGFR mutations, ALK fusions, ROS1 fusions, BRAF V600E mutations, RET fusions, MET amplification, MET exon-14 skipping variants, and ERBB2 (HER2) mutations, are consistently tested. This discrepancy is often due to the unavailability of tumor tissue and the rapid deterioration of a patient’s condition (20). As a result, patients with early mutations often initiate chemotherapy regimens (accounting for 19% of cases) instead of receiving targeted therapies at the outset of their disease (21).
Recent advancements in the identification of predictive biomarkers using models capable of providing a broader genomic view have yielded promising results in the management of NSCLC patients (22). For instance, the utilization of extensive DNA sequencing via liquid biopsy has proven as effective as direct tumor tissue analysis, offering a non-invasive method that exhibits high sensitivity for both NSCLC and solid tumors (23). Full cell-free DNA (cfDNA) analysis through NGS has demonstrated high specificity and strong concordance with tissue-based genotyping. In fact, the use of liquid biopsy, involving a 63-gene sequencing panel via NGS, as compared to real-world tissue analysis, offers sensitivity of 85% and specificity of 99.9% (24-26).
Our study stands out as the initial descriptive and analytical research carried out within the Mexican population to correlate specific molecular mutations in NSCLC patients. We employed NGS using a 62-gene panel to analyze liquid biopsy samples and explore their impact on both PFS and OS.
To describe the clinical characteristics of the entire population with the identified genetic mutations and the therapy received, a heatmap (Figure 2) was created. This heatmap visually represents the population, initially divided into those who received targeted therapy or chemotherapy, along with their respective clinical characteristics and identified genetic mutations.
A prior study conducted on 90 Mexican NSCLC patients using NGS unveiled TP53 (47.8%) and EGFR (36.7%) as the most prevalent mutations (27). Another study conducted on 152 Asian patients, analyzing circulating DNA from lung cancer patients, revealed that the most common alterations in their population were EGFR at 46.05%, ALK at 7.24%, KRAS at 6.58%, and PIK3CA at 6.58%, respectively (28). This contrasts with a study conducted on 229 European patients, in which the most common alterations were TP53 at 47.6%, DNMT3A at 33.2%, EGFR at 20.1%, and KRAS at 15.7% (29). In our series, TP53 was identified as a sole mutation in 10.3% of patients (13 individuals), while it was found in association with other co-mutations in an additional 51.1% of cases. EGFR (24 cases), KRAS (20 cases), ALK (11 cases), and ROS1 (1 case) represented other common mutations. These results underscore the diversity of mutations present in NSCLC patients within the Mexican population. Nevertheless, our data analysis did not reveal a correlation between these mutations and worse OS or PFS. This can be attributed to the fact that while TP53 mutations are linked to poor prognosis, they do not predict the response to chemotherapy treatment (30,31). These mutations, on the other hand, are predictive of immunotherapy agents. Unfortunately, the majority of the population lacks access to these treatments. Notably, 34 out of the total 65 cases with TP53 mutations had a history of tobacco smoking, aligning with findings in the existing literature (32).
An especially significant contribution of our research is the identification and classification of ESCAT level 1 mutations. In our study population, 32.3% (41 patients) presented an actionable genomic variation at ESCAT level 1. This percentage exceeds the reported 29% of NSCLC patients with potentially treatable mutations in prospective studies (33). This finding holds particular importance, underscoring that even in patients preselected as EGFR-negative, NGS can identify target mutations, guiding treatment decisions effectively.
In our study population, EGFR mutations were the most frequently mutated gene (18.9%), closely followed by KRAS (15.8%). Notably, our population, preselected for EGFR mutations, included a substantial proportion of resistance mutations to TKIs and other less common mutations. In a prior study conducted by our group, we demonstrated that 38.19% of cases in the Mexican population tested positive for EGFR mutations through rtPCR (19). Our results identified EGFR mutations in various forms, including six cases with mutations on exon 21 L858R, two of which had co-mutations with T790M. Additionally, there were 14 cases associated with mutations on exon 19, two cases with insertion mutations on exon 20, one case associated with a mutation in exon 18 and one with an impact on exon 7. Another study involving 153 NSCLC patients with deletion mutations on exon 19 discovered 22 distinct deletion types, indicating that unconventional sensitive mutations often elude detection by conventional studies such as rtPCR. Consequently, these patients may miss the opportunity to receive targeted therapy unless they undergo NGS (34). This scenario applies to a subset of the mutations identified in our patients, as some of these mutations may not meet the criteria for targeted therapies. One notable example is the situation involving exon 7, which does not encode for the tyrosine kinase receptor. Consequently, it becomes imperative to explore alternative treatment modalities. In our series, 13 patients with EGFR mutations became eligible for targeted therapy following NGS analysis. This underscores the utility of NGS in identifying less common sensitive mutations for which target therapies are available. Patients with these mutations can benefit from the latest generation of targeted therapies, such as amivantamab for insertion mutations on exon 20, where no treatment options were previously available, or its combination with lazertinib, which is effective for EGFR-MET mutations (35). The presence of BRAF V600E mutation, an uncommon mutational variant found in 1–4% of pulmonary adenocarcinomas, is notable as it shows excellent responses to combination treatments with BRAF and MEK inhibitors, even being reported in studies involving isolated circulating tumor cells (36).
Among our study population, 15.8% had KRAS mutations, with 1.57% of these cases involving the p.G12C mutation. These findings diverge from prior studies, where the p.G12C mutation occurred in 13% of KRAS cases. This discrepancy can be attributed to the small sample size and racial distinctions. Among these patients, five cases had the p.G12C mutation, while eight cases had co-mutations with TP53. In an Italian study involving 194 NSCLC patients, NGS using the SiRe panel identified molecular alterations, with 5% of their cases exhibiting KRAS mutations and no relevant additional genes reported. Remarkably, the p.G12C alteration was found in 36% of their population (37). Other studies report KRAS mutations in 25% of NSCLC patients, with the p.G12C alteration occurring in 13%, highlighting the importance of identifying these mutations, as they respond well to target therapies, offering better disease control rates (80.6%) and prolonged PFS (38).
Our study underscores that utilizing NGS as a method to guide treatment in the Mexican population is a pivotal factor in improving patient survival. The need for new diagnostic tools with high sensitivity and specificity is paramount in identifying genetic mutations that enable patients to receive targeted therapies early in the course of their disease, potentially enhancing both OS and PFS. In developing countries such as Mexico, the transition to employing these novel technologies to guide treatment is an essential step towards providing appropriate and individualized care.
Conclusions
Massive DNA sequencing conducted in the Mexican population reveals that 32.3% of patients who tested negative for an EGFR mutation via rtPCR actually possess target mutations. This finding implies that these patients can initiate targeted therapy at an early stage of their disease, which is associated with prolonged survival. The implementation of massive DNA sequencing is proven to elevate survival rates among patients with non-small cell lung carcinoma. Therefore, the adoption of this technology is essential for guiding the treatment of lung cancer.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1029/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1029/dss
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1029/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1029/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Institutional Ethics Board Committee “Comité de Ética en Investigación Instituto Nacional de Enfermedades Respiratorias” (E08-19) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- SEER. Explorer: An interactive website for SEER cancer statistics. Surveillance Research Program, National Cancer Institute. 2021.
- Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature 2001;409:860-921. [Crossref] [PubMed]
- Alekseyev YO, Fazeli R, Yang S, et al. A Next-Generation Sequencing Primer-How Does It Work and What Can It Do? Acad Pathol 2018;5:2374289518766521. [Crossref] [PubMed]
- Pervez MT, Hasnain MJU, Abbas SH, et al. A Comprehensive Review of Performance of Next-Generation Sequencing Platforms. Biomed Res Int 2022;2022:3457806. [Crossref] [PubMed]
- Morey M, Fernández-Marmiesse A, Castiñeiras D, et al. A glimpse into past, present, and future DNA sequencing. Mol Genet Metab 2013;110:3-24. [Crossref] [PubMed]
- Moorcraft SY, Gonzalez D, Walker BA. Understanding next generation sequencing in oncology: A guide for oncologists. Crit Rev Oncol Hematol 2015;96:463-74. [Crossref] [PubMed]
- Colomer R, Miranda J, Romero-Laorden N, et al. Usefulness and real-world outcomes of next generation sequencing testing in patients with cancer: an observational study on the impact of selection based on clinical judgement. EClinicalMedicine 2023;60:102029. [Crossref] [PubMed]
-
Approves First Liquid Biopsy Next-Generation Sequencing Companion Diagnostic Test FDA 2020 . Available online: https://www.fda.gov/news-events/press-announcements/fda-approves-first-liquid-biopsy-next-generation-sequencing-companion-diagnostic-test - Tan AC, Lai GGY, Tan GS, et al. Utility of incorporating next-generation sequencing (NGS) in an Asian non-small cell lung cancer (NSCLC) population: Incremental yield of actionable alterations and cost-effectiveness analysis. Lung Cancer 2020;139:207-15. [Crossref] [PubMed]
- Pisapia P, Costa JL, Pepe F, et al. Next generation sequencing for liquid biopsy based testing in non-small cell lung cancer in 2021. Crit Rev Oncol Hematol 2021;161:103311. [Crossref] [PubMed]
- Rolfo C, Cardona AF, Cristofanilli M, et al. Challenges and opportunities of cfDNA analysis implementation in clinical practice: Perspective of the International Society of Liquid Biopsy (ISLB). Crit Rev Oncol Hematol 2020;151:102978. [Crossref] [PubMed]
- Caputo V, Ciardiello F, Corte CMD, et al. Diagnostic value of liquid biopsy in the era of precision medicine: 10 years of clinical evidence in cancer. Explor Target Antitumor Ther 2023;4:102-38. [Crossref] [PubMed]
- Chen M, Zhao H. Next-generation sequencing in liquid biopsy: cancer screening and early detection. Hum Genomics 2019;13:34. [Crossref] [PubMed]
- Song X, Wang Z. Clinical efficacy evaluation of tyrosine kinase inhibitors for non-adenocarcinoma lung cancer patients harboring EGFR-sensitizing mutations. Onco Targets Ther 2017;10:3119-22. [Crossref] [PubMed]
- Mayo-de-Las-Casas C, Jordana-Ariza N, Garzón-Ibañez M, et al. Large scale, prospective screening of EGFR mutations in the blood of advanced NSCLC patients to guide treatment decisions. Ann Oncol 2017;28:2248-55. [Crossref] [PubMed]
- Rolfo C, Mack PC, Scagliotti GV, et al. Liquid Biopsy for Advanced Non-Small Cell Lung Cancer (NSCLC): A Statement Paper from the IASLC. J Thorac Oncol 2018;13:1248-68. [Crossref] [PubMed]
- Dziadziuszko R, Mok T, Peters S, et al. Blood First Assay Screening Trial (BFAST) in Treatment-Naïve Advanced or Metastatic Non-Small Cell Lung Cancer: Initial Results of the Phase 2 ALK-Positive Cohort. J Thorac Oncol 2021;16:2040-50. [Crossref] [PubMed]
- Alatorre-Alexander JA, Santillán-Doherty P, Martínez-Barrera LM, et al. Descripción clínico-epidemiológica y molecular del cáncer de pulmón en un centro de referencia nacional. NCT Neumol y Cirugía Tórax 2019;78:356-62.
- Leighl NB, Page RD, Raymond VM, et al. Clinical Utility of Comprehensive Cell-free DNA Analysis to Identify Genomic Biomarkers in Patients with Newly Diagnosed Metastatic Non-small Cell Lung Cancer. Clin Cancer Res 2019;25:4691-700. [Crossref] [PubMed]
- Lim C, Tsao MS, Le LW, et al. Biomarker testing and time to treatment decision in patients with advanced non-small-cell lung cancer. Ann Oncol 2015;26:1415-21. [Crossref] [PubMed]
- Zill OA, Banks KC, Fairclough SR, et al. The Landscape of Actionable Genomic Alterations in Cell-Free Circulating Tumor DNA from 21,807 Advanced Cancer Patients. Clin Cancer Res 2018;24:3528-38. [Crossref] [PubMed]
- Cheng J, Cao Y, MacLeay A, et al. Clinical Validation of a Cell-Free DNA Gene Panel. J Mol Diagn 2019;21:632-45. [Crossref] [PubMed]
- Palmero R, Taus A, Viteri S, et al. P2.03-02 Cell-Free DNA (cfDNA) Testing in Lung Adenocarcinoma (LUAC) Patients: Spanish Lung Liquid Versus Invasive Biopsy Program (SLLIP). J Thorac Oncol 2018;13:S716-7.
- Clark TA, Chung JH, Kennedy M, et al. Analytical Validation of a Hybrid Capture-Based Next-Generation Sequencing Clinical Assay for Genomic Profiling of Cell-Free Circulating Tumor DNA. J Mol Diagn 2018;20:686-702. [Crossref] [PubMed]
- Legras A, Barritault M, Tallet A, et al. Validity of Targeted Next-Generation Sequencing in Routine Care for Identifying Clinically Relevant Molecular Profiles in Non-Small-Cell Lung Cancer: Results of a 2-Year Experience on 1343 Samples. J Mol Diagn 2018;20:550-64. [Crossref] [PubMed]
- Hernández-Pedro N, Soca-Chafre G, Alaez-Versón C, et al. Mutational profile by targeted next generation sequencing of non-small cell lung cancer in the Mexican population. Salud Publica Mex 2019;61:308-17. [Crossref] [PubMed]
- Zhao S. Biomed Res Int 2021;2021:8817898. [Crossref] [PubMed]
- Porta C, Pradelli L, Sicari E, et al. Liquid biopsy comprehensive genomic profiling of lung cancer in the Italian population: A real-world experience. Lung Cancer 2023;185:107359. [Crossref] [PubMed]
- Mehta A, Vasudevan S, Sharma SK, et al. Biomarker testing for advanced lung cancer by next-generation sequencing; a valid method to achieve a comprehensive glimpse at mutational landscape. Appl Cancer Res 2020;40:4.
- Molina-Vila MA, Bertran-Alamillo J, Gascó A, et al. Nondisruptive p53 mutations are associated with shorter survival in patients with advanced non-small cell lung cancer. Clin Cancer Res 2014;20:4647-59. [Crossref] [PubMed]
- Halvorsen AR, Silwal-Pandit L, Meza-Zepeda LA, et al. TP53 Mutation Spectrum in Smokers and Never Smoking Lung Cancer Patients. Front Genet 2016;7:85. [Crossref] [PubMed]
- Lee DH, Subbiah V, Gainor JF, et al. Treatment with pralsetinib (formerly BLU-667), a potent and selective RET inhibitor, provides rapid clearance of ctDNA in patients with RET-altered non-small cell lung cancer (NSCLC) and medullary thyroid cancer (MTC). Mini Oral Session Presented at: ESMO Asia Congress 2019. Ann Oncol 2019;30:ix122-30.
- Oskina N, Oscorbin I, Khrapov E, et al. Highly Sensitive and Reliable Detection of EGFR Exon 19 Deletions by Droplet Digital Polymerase Chain Reaction. Mol Diagn Ther 2017;21:555-62. [Crossref] [PubMed]
- Guo MZ, Marrone KA, Spira A, et al. Amivantamab: A Potent Novel EGFR/c-MET Bispecific Antibody Therapy for. touchREVIEWS in Oncology & Haematology 2021;17:42-7.
- Mastromarino MG, Parini S, Azzolina D, et al. Liquid Biopsy Detecting Circulating Tumor Cells in Patients with Non-Small Cell Lung Cancer: Preliminary Results of a Pilot Study. Biomedicines 2023;11:153. [Crossref] [PubMed]
- Nacchio M, Sgariglia R, Gristina V, et al. KRAS mutations testing in non-small cell lung cancer: the role of Liquid biopsy in the basal setting. J Thorac Dis 2020;12:3836-43. [Crossref] [PubMed]
- Skoulidis F, Li BT, Dy GK, et al. Sotorasib for Lung Cancers with KRAS p.G12C Mutation. N Engl J Med 2021;384:2371-81. [Crossref] [PubMed]