
Increased LOXL2 expression is related to poor prognosis in lung squamous cell carcinoma
Highlight box
Key findings
• Increased lysyl oxidase-like protein 2 (LOXL2) was related to poor survival in lung squamous cell carcinoma (LUSC). High LOXL2 expression was an independent prognostic factor for poor survival.
• Inhibition of LOXL2 suppressed proliferation, migration and invasion in LUSC cell lines.
What is known and what is new?
• LOXL was involved in the process of cancer development. LOXL2 and LOXL4 is upregulated in some tumors and related to more aggressive biological behavior. However, the expression pattern and prognostic value of LOXL family members in LUSC remains unknown.
• We found that LOXL1 and LOXL2 expression was upregulated in LUSC tissues and showed high diagnostic power in LUSC patients. High LOXL2 expression was an independent prognostic factor for poor survival.
What is the implication, and what should change now?
• LOXL2 may be a potential prognostic biomarker and therapeutic target in LUSC. But the molecular mechanism of LOXL2 should be further validated by molecular biology experiments. The diagnostic and prognostic values of LOXL2 in LUSC should be further validated clinically.
Introduction
Non-small cell lung cancer (NSCLC) accounts for 85% of lung cancers. Lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) are the main pathological types of NSCLC (1). For early-stage NSCLC patients, surgical resection is the main therapy. However, surgical resection is not suitable for advanced-stage patients. For these patients, chemotherapy, targeted therapy or immunotherapy are the recommended treatment methods (2). Targeted therapy has made great achievements in LUAD. Some molecular targets have been identified and approved for clinical use, including epidermal growth factor receptor (EGFR), anaplastic lymphoma kinase (ALK) and ROS proto-oncogene 1 (ROS1) rearrangement (3). Drugs targeting these molecules have shown good efficacy in LUAD but not in squamous cell carcinoma. Immune checkpoint inhibitors in combination with chemotherapy remain the first-line therapy for advanced LUSC patients (4). Although this treatment strategy significantly improves the prognosis of LUSC, some serious side effects have been reported (5,6). Investigation of novel diagnostic and prognostic biomarkers may contribute to personalized treatment and help to improve the survival of LUSC patients.
Lysyl oxidase (LOX) is a copper-dependent monoamine oxidase in the extracellular matrix (ECM). There are four LOX isozymes in humans, namely, lysyl oxidase-like protein 1/2/3/4 (LOXL1/2/3/4) (7). It was reported that LOXL was involved in the process of cancer development (8). LOXL2 is upregulated in breast cancer, gastric cancer and cervical cancer and is related to more aggressive biological behavior (9,10). LOXL4 is overexpressed in head and neck squamous cell carcinoma (HNSCC) and promotes resistance to chemotherapy and radiotherapy (8,11). In LUAD, overexpression of LOX is associated with poor prognosis and invasion (12). Targeting EGFR/LOX pathway inhibits metastasis in LUAD (13). However, the expression pattern and prognostic value of LOXL family members in LUSC remains unknown.
High-throughput sequencing and genomic microarray technology combined with bioinformatic analysis provide a powerful tool for the discovery of novel biomarkers. In the current study, we investigated the expression pattern and prognostic value of the LOXL family in LUSC using bioinformatic methods. Then, we further showed that inhibition of LOXL2 suppressed the proliferation, migration, and invasion of LUSC cells. We present this article in accordance with the MDAR reporting checklist (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1848/rc).
Methods
Datasets and clinical information
LOXL expression information for 49 normal lung tissues and 502 LUSC tissues was downloaded from The Cancer Genome Atlas (TCGA)-LUSC. Clinical data of LUSC patients were also downloaded. The analysis was performed using R v3.6.6 software. The expression difference of the LOXL family was visualized by dot graphs. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Gene set enrichment analysis (GSEA)
Patients were classified as LOXL-high and LOXL-low phenotypes based on the median expression level of LOXL. GSEA was performed to assess the potential pathways related to LOXL in LUSC. GSEA v4.1.0 software was used to conduct GSEA. “c2.cp.kegg.v6.2.symbols.gmt” was used as a reference gene set. The number of gene set permutations was 1,000. The enriched gene sets were considered significant if the false discovery rate (FDR) Q-value was less than 0.05.
Cell culture
The H226 and H520 cell lines were purchased from Procell Co., Ltd. (Wuhan, China, H226, cat# CL-0396 and H520, cat# CL-0402). Cells were cultured in RPMI-1640 medium (Gibco, Thermo Fisher Scientific, Inc., MA, USA, cat# 11875119) containing 10% fetal bovine serum (FBS) (Beyotime, Wuhan, China, cat# C0234) and 1% penicillin/streptomycin (Beyotime, cat# C0222). Cells were incubated in a 5% CO2 humidified incubator at 37 ℃.
Construction of LOXL2 knockdown (KD) cell lines
Cells (5×105 cell/well) were seeded into six-well plates the day before transfection. LOXL2 siRNA and the negative control (NC) siRNA were purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, China, cat# SIGS0006971-1). A mixture composed of 125 µL OPTI-MEM (Thermo Fisher Scientific, Inc., cat# 31985062), 4 µL LipoRNAiTM (Beyotime, cat# C0535) and 100 pmol siRNA was prepared and incubated in a six-well dish for 20 min at 37 ℃. The culture medium in the wells was replaced with fresh medium, and 125 µL of the above mixture was added to each well. Then, the cells were incubated for 48 h.
Cell proliferation assay
Cell proliferation was tested using the Cell Counting Kit-8 (CCK-8) kit (Beyotime, cat# C0038). Cells (2,000–3,000 cells/well with a volume of 100 µL) were seeded into a 96-well plate and treated with a LOXL2 inhibitor (LOXL2-IN-1, Baiaolaibo, Beijing, China, cat# M03295) or vehicle [dimethyl sulfoxide (DSMO), Beyotime, cat# ST038] the next day (or incubated if no treatment was needed). Then, the cells were incubated for 24, 48, and 72 hours. The medium was removed, and 100 µL fresh medium with 10 µL CCK-8 solution was added to each well. The cells were incubated in the dark for 1 hour. After that, the optical density (OD) value at 450 nm was measured with a microplate reader. The assay was repeated three times.
Wound healing assay
Cells were seeded into a 12-well plate and grown to approximately 95% confluency. A scratch wound was created in each well. Then, the cells were washed, and fresh media (with LOXL2 inhibitor or vehicle in the LOXL2 inhibitor and vehicle group) was added. After incubation for 24 hours, the wounds were imaged and measured using ImageJ software. The assay was repeated three times.
Cell invasion assay
Cell invasion was determined using a Transwell with 8 µm pores (Corning, USA). The Matrigel matrix was removed from −80 ℃ and defrosted at 4 ℃ on ice. The matrix was swirled to distribute the material evenly. Then, the Matrigel matrix was diluted to a concentration of 0.3 mg/mL using cold medium not containing serum. The diluted Matrigel matrix was added to a 24-well plate (100 µL/well) in the Transwell insert. Then, the 24-well plates were incubated for 1–2 hours at 37 ℃. After that, the remaining liquid was removed carefully. Cells (2.5×104 cells) were seeded into the upper chamber of a Transwell in 100 µL of serum-free medium (with LOXL2 inhibitor or vehicle in the LOXL2 inhibitor and vehicle group). Medium (700 µL) containing 10% FBS was added to the bottom chamber. The cells were incubated for 12 hours. Nonmigrated cells were removed, and the migrated cells were fixed with 4% paraformaldehyde for 30 minutes and stained with crystal violet. The migrated cells were counted (×100 magnification). The assay was repeated three times.
Western blotting
The medium was removed, and the cells were washed with PBS. Cells were lysed using RIPA buffer (Beyotime, cat# P0013B). The BCA Protein Assay Kit (Beyotime, cat# P0010S) was used to determine the protein concentration. The protein (40 µg/sample) was loaded and separated by a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred to a poly vinylidene fluoride (PVDF) membrane. After blotting with 5% bovine serum albumin (BSA), the membrane was incubated with rabbit anti-LOXL2 antibody (1:1,000, Abcam, MA, USA, cat# ab179810) and mouse anti-β-actin antibody (1:5,000, Beyotime, cat# AF5001) at 4 ℃ overnight. Then, the membrane was washed with tris buffered saline (TBS) and incubated with goat anti-rabbit (1:2,000, Beyotime, cat# A0208) or goat anti-mouse antibody (1:2,000, Beyotime, cat# A0216) for 1 hour at room temperature. The bands were visualized using enhanced chemiluminescence (ECL) Plus reagent (Beyotime, cat# P0018S). The assay was repeated three times.
Statistical analysis
Statistical analyses were performed using R v3.6.3 software. The Wilcoxon test was used to compare the LOXL expression level between normal and tumor tissues. The association between LOXL expression and clinical characteristics was analyzed by logistic regression analysis. Receiver operator characteristic (ROC) curves were used to detect the diagnostic power of LOXL1 and LOXL2 in LUSC. Correlations between clinical characteristics and survival were analyzed by univariate and multivariate Cox regression analyses. If the P value in the univariate analysis was less than 0.1, the variable would be included in the multivariate Cox regression analysis. Other results were analyzed using Student’s t-test unless otherwise stated. The results were considered statistically significant if P<0.05.
Results
LOXL1 and LOXL2 were upregulated in LUSC tissues
LOXL1/2/3/4 expression levels in 502 LUSC tissues and 49 normal tissues were compared. Three hundred and seventy-one males and 472 smokers were enrolled in the analyses. Unpaired comparisons showed that LOXL1 and LOXL2 expression was upregulated in LUSC tissues (P<0.001). However, the expression of LOXL3 and LOXL4 showed no significant difference between normal and tumor tissues (Figure 1A). The expression of LOXL1/2/3/4 in 49 pairs of cancer and adjacent noncancerous tissues was also analyzed. As shown in Figure 1B, LOXL1 and LOXL2 were significantly increased in LUSC tissues (P<0.001).

Correlation of LOXL1 and LOXL2 with clinical characteristics
The expression of LOXL1 and LOXL2 in patients with different clinical characteristics was further analyzed. LOXL1 expression was higher in patients older than 65 years (P<0.05), while LOXL2 expression showed no difference between different age subgroups (Figure 1C). LOXL1 and LOXL2 expression in patients with different gender, smoking statuses, T stages, N stages, M stages and pathologic stages did not show significant differences (Figure 1D-1I). The relationship between LOXL1 and LOXL2 expression and clinical characteristics was further estimated by logistic regression analysis. The results showed that both LOXL1 and LOXL2 were not related to age, gender, smoking status, T stage, N stage or M stage (Table 1).
Table 1
| Characteristics | Total, N | OR (95% CI) | P value |
|---|---|---|---|
| LOXL1 | |||
| Age (>65 vs. ≤65 years) | 493 | 1.274 (0.887–1.834) | 0.191 |
| Gender (male vs. female) | 502 | 1.156 (0.776–1.724) | 0.477 |
| Smoker (yes vs. no) | 490 | 2.017 (0.770–5.876) | 0.168 |
| T stage (T2&T3&T4 vs. T1) | 502 | 0.956 (0.629–1.452) | 0.831 |
| N stage (N1&N2&N3 vs. N0) | 496 | 0.858 (0.593–1.239) | 0.414 |
| M stage (M1 vs. M0) | 419 | 1.246 (0.271–6.389) | 0.775 |
| LOXL2 | |||
| Age (>65 vs. ≤65 years) | 493 | 1.347 (0.937–1.940) | 0.108 |
| Gender (male vs. female) | 502 | 1.309 (0.878–1.957) | 0.187 |
| Smoker (yes vs. no) | 490 | 1.585 (0.613–4.372) | 0.350 |
| T stage (T2&T3&T4 vs. T1) | 502 | 1.441 (0.947–2.202) | 0.089 |
| N stage (N1&N2&N3 vs. N0) | 496 | 0.763 (0.527–1.103) | 0.152 |
| M stage (M1 vs. M0) | 419 | 6.118 (1.033–116.076) | 0.095 |
LOXL1, lysyl oxidase-like protein 1; LOXL2, lysyl oxidase-like protein 2; OR, odds ratio; CI, confidence interval; T, tumor; N, node; M, metastasis.
Diagnostic power of LOXL1 and LOXL2 in LUSC patients
To assess the diagnostic power of LOXL1 and LOXL2 in LUSC, ROC curves were plotted, and areas under the curves (AUCs) were calculated. LOXL1 showed a sensitivity of 0.631 and a specificity of 0.791. The AUC of LOXL1 was 0.784. LOXL2 showed a sensitivity of 0.735 and a specificity of 0.703. The AUC of LOXL2 was 0.751 (Figure 1J). The results indicated that LOXL1 and LOXL2 had high diagnostic power in LUSC patients.
High expression of LOXL2 was related to poor survival in LUSC
Patients were classified into high and low-expression groups based on the median expression levels of LOXL1 and LOXL2. The survival of patients with high and low LOXL1/2 expression was compared by the Kaplan-Meier method. The overall survival (OS) and progression-free survival (PFS) of patients in the LOXL1 high and low-expression groups were not significantly different (Figure 2A,2B). Patients with high LOXL2 expression showed worse OS [hazard ratio (HR) =1.38; 95% confidence interval (CI): 1.05–1.82; P=0.019] and PFS (HR =1.50; 95% CI: 1.08–2.08; P=0.015) than patients with low LOXL2 expression (Figure 2C,2D). The median OS of patients with high and low LOXL2 expression was 36.9 months (95% CI: 32.5–61.9) and 65.8 months (95% CI: 54.7–80.3), respectively. The median PFS of patients with high and low LOXL2 expression was 55.2 months (95% CI: 48.3–76.4) and 102.7 months (95% CI: 76.8–not reach), respectively. The results indicated that high expression of LOXL2 was related to poor survival in LUSC.

The prognostic power of LOXL2 in LUSC
Univariate and multivariate Cox regression analyses were performed to estimate the correlation of LOXL2 expression with the prognosis of LUSC. Univariate analysis indicated that high LOXL2 expression (HR =1.383; 95% CI: 1.054–1.815; P=0.019) and M1 stage (HR =3.112; 95% CI: 1.272–7.616; P=0.013) were related to poor survival. Multivariate analysis showed that age older than 65 years (HR =1.380; 95% CI: 1.003–1.898; P=0.048), M1 stage (HR =2.516; 95% CI: 1.018–6.222; P=0.046) and high LOXL2 expression (HR =1.411; 95% CI: 1.042–1.910; P=0.026) were independent prognostic factors for LUSC (Table 2). ROCs were plotted, and AUCs were calculated to estimate the prognostic power of LOXL2 for OS and PFS. The predictive power of LOXL2 for OS is shown in Figure 2E. The AUCs of the ROC curves for 1-, 3- and 5-year OS were 0.554, 0.583 and 0.562, respectively. The predictive power of LOXL2 for PFS is shown in Figure 2F. The AUCs of the ROC curves for 1-, 3- and 5-year PFS were 0.558, 0.590 and 0.575, respectively. The results suggested that high LOXL2 expression had a mild predictive power for poor clinical outcome in LUSC.
Table 2
| Characteristics | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | ||
| Age | |||||
| ≤65 years | Reference | Reference | |||
| >65 years | 1.279 (0.960–1.704) | 0.093 | 1.380 (1.003–1.898) | 0.048 | |
| Gender | |||||
| Female | Reference | Reference | |||
| Male | 1.211 (0.879–1.669) | 0.241 | – | – | |
| T stage | |||||
| T1 | Reference | Reference | |||
| T2&T3&T4 | 1.377 (0.984–1.926) | 0.062 | 1.177 (0.816–1.698) | 0.383 | |
| N stage | |||||
| N0 | Reference | Reference | |||
| N1&N2&N3 | 1.151 (0.869–1.523) | 0.327 | – | – | |
| M stage | |||||
| M0 | Reference | Reference | |||
| M1 | 3.112 (1.272–7.616) | 0.013 | 2.516 (1.018–6.222) | 0.046 | |
| LOXL2 | |||||
| Low | Reference | Reference | |||
| High | 1.383 (1.054–1.815) | 0.019 | 1.411 (1.042–1.910) | 0.026 | |
LOXL2, lysyl oxidase-like protein 2; HR, hazard ratio; CI, confidence interval; T, tumor; N, node; M, metastasis.
Inhibition of LOXL2 suppressed cell proliferation, migration, and invasion of LUSC cell lines
We knocked down LOXL2 expression in the LUSC cell lines H226 and H520 by transfection of siRNA targeting LOXL2. Western blotting showed that LOXL2 was significantly decreased in the LOXL2 group compared to the control group and NC group (Figure 3A-3D). LOXL2 expression was decreased in the LOXL2 KD groups and did not show a significant difference in the NC groups. The results indicated that LOXL2 was knocked down in the LOXL2 KD groups of H226 and H520 cell lines. Then, we compared the differences in proliferation, migration, and invasion abilities between cells with different LOXL2 expression levels. We used CCK8 assays to detect cell proliferation. In comparison to that in the NC groups, cell proliferation was inhibited in the LXOL2 KD groups (Figure 3E,3F). Wound healing assays were used to estimate cell migration. A larger migration distance suggests a stronger migration ability. Migration distances in the LOXL2 KD groups were significantly smaller than those in the NC groups (Figure 3G,3H). Transwell assays were used to determine cell invasion ability. The more cells that migrated from the upper chamber to the bottom chamber of the Transwell, the higher the invasion capacity was. The results showed that the cell count in the bottom chamber in LOXL2 KD cells was significantly lower than that in the NC groups (Figure 3I,3J). To further validate the influence of LOXL2 on cell behaviors, we used an LXOL2 inhibitor (LOXL2-IN-1, 60 nM) to treat LUSC cells. The vehicle groups were used as control groups. The results showed that compared with the vehicle groups, the cell proliferation, migration and invasion of the LOXL2 inhibitor groups were obviously suppressed (Figure 3E-3J). The results indicated that inhibition of LOXL2 (knocking down LOXL2 expression or treatment with a LOXL2 inhibitor) suppressed the proliferation, migration, and invasion of LUSC cell lines.

Identification of LOXL2-related pathways
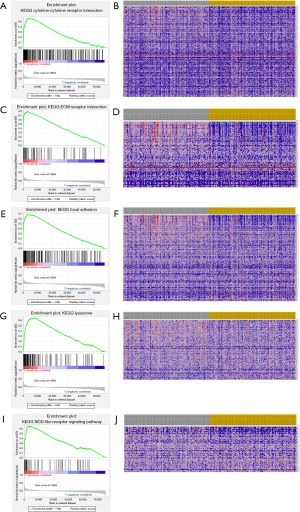
Patients were classified into the LOXL2-high phenotype and LOXL2-low phenotype. GSEA was conducted to screen LOXL2-related pathways. The cytokine-cytokine receptor interaction pathway, ECM receptor interaction pathway, focal adhesion (FA) pathway, lysosome pathway, and NOD-like receptor signaling pathway were enriched in the LOXL2-high phenotype (Figure 4).
Discussion
In the current study, we found that LOXL1 and LOXL2 expression was upregulated in LUSC tissues and showed high diagnostic power in LUSC patients. High LOXL2 expression was an independent prognostic factor for poor survival.
Lysyl oxidate-like (LOXL) protein plays an important role in catalyzing the covalent cross-linking of collagens and elastin in the ECM. There are four types of LOXL, namely, LOXL1, LOXL2, LOXL3 and LOXL4 (14). LOXL1/2/3/4 are involved in tumor development. LOXL1 is increased and promotes tumorigenesis in LUAD and prostate cancer (15,16). LOXL2 expression is related to poor prognosis in colon cancer, esophageal cancer, and breast cancer (17). LOXL3 interacts with Snail homolog 1 (SNAIL) and promotes cell proliferation and tumor invasion and metastasis in pancreatic ductal adenocarcinoma and gastric cancer (18). LOXL4 promotes metastasis of liver cancer and colorectal cancer (19,20). However, the role of LOXL in LUSC remains unknown. It has been pointed out that overexpression of integrin α11 increased LOXL1 expression in NSCLC and promotes tumor growth and progression. LOXL1 induces NSCLC cell invasion by affecting collagen matrix remodeling (21). In addition, LOXL1 is associated with chemotherapy resistance in NSCLC (21). Thus, we performed the current research to investigate the expression pattern and role of LOXL in LUSC.
First, we compared the expression levels of LOXL1/2/3/4 in LUSC tissues and noncancerous tissues. We found that LOXL1 and LOXL2 were upregulated in LUSC. However, neither LOXL1 nor LOXL2 expression was related to T stage, N stage, M stage or pathological stage. The results suggested that upregulation of LOXL1 and LOXL2 may be involved in tumorigenesis rather than tumor progression. We further plotted the ROCs to assess the diagnostic power of LOXL1 and LOXL2 in LUSC. The results showed that both LOXL1 and LOXL2 showed high diagnostic power in LUSC.
Second, we performed survival analysis and Cox regression analysis to estimate the correlation of LOXL1 and LOXL2 with LUSC patient survival. The results indicated that a high LOXL2 expression level was related to poor survival, while the LOXL1 expression level was not related to survival. Univariate and multivariate Cox regression showed that LOXL2 was an independent prognostic biomarker for poor survival. It was reported in 2003 for the first time that LOXL2 promoted tumor fibrosis and was related to more aggressive behavior in breast cancer (22). Since then, an increasing number of studies on LOXL2 and cancer have been performed. High expression of LOXL2 was reported to be associated with poor survival in HNSCC, LUAD, esophageal cancer, colon cancer, liver cancer and cervical cancer (22). In this study, we found that LOXL2 was also increased in LUSC and related to poor survival. To further investigate the effect of LOXL2 on the biological behavior of LUSC cells, we knocked down LOXL2 expression in LUSC cell lines and compared the proliferation, migration and invasion abilities of cells with different LOXL2 expression levels. We also explored the effect of an LXOL2 inhibitor on LUSC cells. The results indicated that inhibition of LOXL2, either by knocking down LOXL2 expression or by a LOXL2 inhibitor, suppressed the proliferation, migration and invasion abilities of LUSC cells. The results also supported that LOXL2 was a prognostic biomarker for poor survival. However, in human body, in addition to oncogenes and oncogenes, tumor development is affected by many other factors, such as the immune system, lipid metabolism, tumor microenvironment and so on. The in vitro experiments only examined the effects of LOXL1 and LOXL2 on tumor cells, and did not examine the effects of other factors.
LOXL2 regulates ECM remodeling and promotes tumor invasion. A recent study showed that LOXL2 can also promote tumor progression in esophageal cancer by activating glycolytic enzymes such as aldolase A (23). Inhibiting LOXL2 using small molecule inhibitors restrains malignant transformation of cervical cancer cells by suppressing epithelial-mesenchymal transition (EMT) (24). To perform a preliminary exploration of the mechanism of LOXL2, we used GSEA to screen LXOL2-associated signaling pathways. We found that the ECM receptor interaction pathway and FA pathway were enriched in the LOXL2-high phenotype. FA is a subcellular structure that strongly adheres to the ECM and acts as a scaffold for many integrin- or mechanical force-related signaling pathways (25). It was reported that KD of LOXL2 suppressed FA formation in renal cell carcinoma (RCC) (26). Another study indicated that LOXL2 activated fibroblasts by activating FA kinase and promoted cancer progression (27). The results suggested that LOXL2 may promote tumorigenesis through FA-related processes in LUSC.
However, some limitations of this study should be noted. First, the number of cancer tissues was much higher than the number of noncancerous tissues in the TCGA database. Second, only the mRNA expression of the tissue was analyzed. The protein expression level of LOXL was unknown. Finally, the potential pathways related to LOXL2 were only analyzed by bioinformatic analysis. The molecular mechanism should be further validated by molecular biology experiments.
Conclusions
In conclusion, we found that LOXL1 and LOXL2 were upregulated in LUSC and showed high diagnostic power. Overexpression of LOXL2 was related to poor survival and was an independent prognostic biomarker in LUSC. LOXL2 may be a potential prognostic biomarker and therapeutic target in LUSC. But the molecular mechanism of LOXL2 should be further validated by molecular biology experiments. The diagnostic and prognostic values of LOXL2 in LUSC should be further validated clinically.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1848/rc
Data Sharing Statement: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1848/dss
Peer Review File: Available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1848/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-23-1848/coif). C.Z. is from Shenzhen Yuce Biotechnology Co. Ltd. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Gao M, Kong W, Huang Z, et al. Identification of Key Genes Related to Lung Squamous Cell Carcinoma Using Bioinformatics Analysis. Int J Mol Sci 2020;21:2994. [Crossref] [PubMed]
- Ettinger DS, Wood DE, Aggarwal C, et al. NCCN Guidelines Insights: Non-Small Cell Lung Cancer, Version 1.2020. J Natl Compr Canc Netw 2019;17:1464-72. [Crossref] [PubMed]
- Oberndorfer F, Müllauer L. Molecular pathology of lung cancer: current status and perspectives. Curr Opin Oncol 2018;30:69-76. [Crossref] [PubMed]
- Fan FS, Yang CF, Chang CL. Nivolumab plus Carboplatin and Paclitaxel as the First-line Therapy for Advanced Squamous Cell Carcinoma of the Lung with Strong Programmed Death-ligand 1 Expression: A Case Report. Cureus 2019;11:e5881. [Crossref] [PubMed]
- Paz-Ares L, Luft A, Vicente D, et al. Pembrolizumab plus Chemotherapy for Squamous Non-Small-Cell Lung Cancer. N Engl J Med 2018;379:2040-51. [Crossref] [PubMed]
- Matsubara T, Uchi H, Haratake N, et al. Acute Generalized Exanthematous Pustulosis Caused by the Combination of Pembrolizumab Plus Chemotherapy in a Patient With Squamous-Cell Carcinoma. Clin Lung Cancer 2020;21:e54-6. [Crossref] [PubMed]
- Asuncion L, Fogelgren B, Fong KS, et al. A novel human lysyl oxidase-like gene (LOXL4) on chromosome 10q24 has an altered scavenger receptor cysteine rich domain. Matrix Biol 2001;20:487-91. [Crossref] [PubMed]
- Barker HE, Cox TR, Erler JT. The rationale for targeting the LOX family in cancer. Nat Rev Cancer 2012;12:540-52. [Crossref] [PubMed]
- Kasashima H, Yashiro M, Okuno T, et al. Significance of the Lysyl Oxidase Members Lysyl Oxidase Like 1, 3, and 4 in Gastric Cancer. Digestion 2018;98:238-48. [Crossref] [PubMed]
- Barker HE, Chang J, Cox TR, et al. LOXL2-mediated matrix remodeling in metastasis and mammary gland involution. Cancer Res 2011;71:1561-72. [Crossref] [PubMed]
- Görögh T, Holtmeier C, Weise JB, et al. Functional analysis of the 5’ flanking domain of the LOXL4 gene in head and neck squamous cell carcinoma cells. Int J Oncol 2008;33:1091-8.
- Wilgus ML, Borczuk AC, Stoopler M, et al. Lysyl oxidase: a lung adenocarcinoma biomarker of invasion and survival. Cancer 2011;117:2186-91. [Crossref] [PubMed]
- Hou X, Du H, Quan X, et al. Silibinin Inhibits NSCLC Metastasis by Targeting the EGFR/LOX Pathway. Front Pharmacol 2018;9:21. [Crossref] [PubMed]
- Vallet SD, Berthollier C, Salza R, et al. The Interactome of Cancer-Related Lysyl Oxidase and Lysyl Oxidase-Like Proteins. Cancers (Basel) 2020;13:71. [Crossref] [PubMed]
- Ji H, Ramsey MR, Hayes DN, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature 2007;448:807-10. [Crossref] [PubMed]
- Nilsson M, Adamo H, Bergh A, et al. Inhibition of Lysyl Oxidase and Lysyl Oxidase-Like Enzymes Has Tumour-Promoting and Tumour-Suppressing Roles in Experimental Prostate Cancer. Sci Rep 2016;6:19608. [Crossref] [PubMed]
- Moon HJ, Finney J, Ronnebaum T, et al. Human lysyl oxidase-like 2. Bioorg Chem 2014;57:231-41. [Crossref] [PubMed]
- Laurentino TS, Soares RDS, Marie SKN, et al. LOXL3 Function Beyond Amino Oxidase and Role in Pathologies, Including Cancer. Int J Mol Sci 2019;20:3587. [Crossref] [PubMed]
- Li R, Wang Y, Zhang X, et al. Exosome-mediated secretion of LOXL4 promotes hepatocellular carcinoma cell invasion and metastasis. Mol Cancer 2019;18:18. [Crossref] [PubMed]
- Palmieri V, Lazaris A, Mayer TZ, et al. Neutrophils expressing lysyl oxidase-like 4 protein are present in colorectal cancer liver metastases resistant to anti-angiogenic therapy. J Pathol 2020;251:213-23. [Crossref] [PubMed]
- Zeltz C, Pasko E, Cox TR, et al. LOXL1 Is Regulated by Integrin α11 and Promotes Non-Small Cell Lung Cancer Tumorigenicity. Cancers (Basel) 2019;11:705. [Crossref] [PubMed]
- Wen B, Xu LY, Li EM. LOXL2 in cancer: regulation, downstream effectors and novel roles. Biochim Biophys Acta Rev Cancer 2020;1874:188435. [Crossref] [PubMed]
- Jiao JW, Zhan XH, Wang JJ, et al. LOXL2-dependent deacetylation of aldolase A induces metabolic reprogramming and tumor progression. Redox Biol 2022;57:102496. [Crossref] [PubMed]
- Peng T, Lin S, Meng Y, et al. LOXL2 small molecule inhibitor restrains malignant transformation of cervical cancer cells by repressing LOXL2-induced epithelial-mesenchymal transition (EMT). Cell Cycle 2022;21:1827-41. [Crossref] [PubMed]
- Shen J, Cao B, Wang Y, et al. Hippo component YAP promotes focal adhesion and tumour aggressiveness via transcriptionally activating THBS1/FAK signalling in breast cancer. J Exp Clin Cancer Res 2018;37:175. [Crossref] [PubMed]
- Hase H, Jingushi K, Ueda Y, et al. LOXL2 status correlates with tumor stage and regulates integrin levels to promote tumor progression in ccRCC. Mol Cancer Res 2014;12:1807-17. [Crossref] [PubMed]
- Correction: Tumor-Secreted LOXL2 Activates Fibroblasts through FAK Signaling. Mol Cancer Res 2019;17:2141. [Crossref] [PubMed]


